['Non-cancer illness code, self-reported',
'Albumin',
'Alkaline phosphatase',
'Alanine aminotransferase',
'Apolipoprotein A',
'Apolipoprotein B',
'Aspartate aminotransferase',
'Urea',
'Calcium',
'Cholesterol',
'Creatinine',
'C-reactive protein',
'Cystatin C',
'Gamma glutamyltransferase',
'Glycated haemoglobin (HbA1c)',
'HDL cholesterol',
'IGF-1',
'Phosphate',
'SHBG',
'Total protein',
'Triglycerides',
'Urate',
'Pulse rate, automated reading',
'Time spent watching television (TV)',
'Morning/evening person (chronotype)',
'Ventricular rate',
'P duration',
'QRS duration',
'Comparative body size at age 10',
'Comparative height size at age 10',
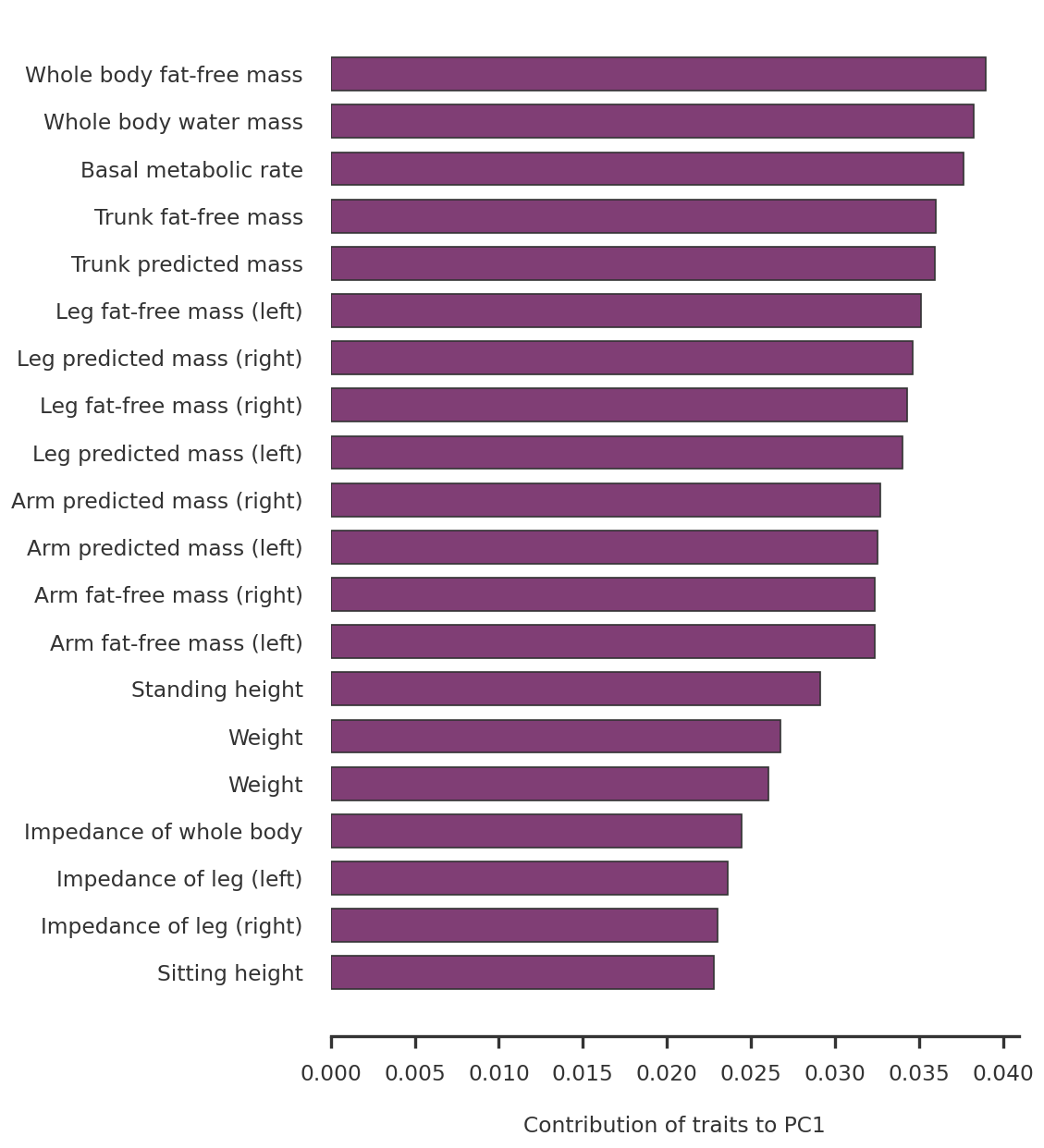
'Sitting height',
'Fluid intelligence score',
'Birth weight',
'Neuroticism score',
'Forced expiratory volume in 1-second (FEV1), Best measure',
'Forced vital capacity (FVC), Best measure',
'Forced expiratory volume in 1-second (FEV1), predicted',
'Forced expiratory volume in 1-second (FEV1), predicted percentage',
'Body mass index (BMI)',
'Weight',
'Age first had sexual intercourse',
'Overall health rating',
'Age hayfever or allergic rhinitis diagnosed by doctor',
'Age asthma diagnosed by doctor',
'Posterior thigh lean muscle volume (right)',
'Posterior thigh lean muscle volume (left)',
'Visceral adipose tissue volume (VAT)',
'Abdominal subcutaneous adipose tissue volume (ASAT)',
'Total trunk fat volume',
'Year ended full time education',
'Minimum carotid IMT (intima-medial thickness) at 120 degrees',
'Mean carotid IMT (intima-medial thickness) at 120 degrees',
'Maximum carotid IMT (intima-medial thickness) at 120 degrees',
'Minimum carotid IMT (intima-medial thickness) at 150 degrees',
'Mean carotid IMT (intima-medial thickness) at 150 degrees',
'Maximum carotid IMT (intima-medial thickness) at 150 degrees',
'Minimum carotid IMT (intima-medial thickness) at 210 degrees',
'Mean carotid IMT (intima-medial thickness) at 210 degrees',
'Maximum carotid IMT (intima-medial thickness) at 210 degrees',
'Minimum carotid IMT (intima-medial thickness) at 240 degrees',
'Mean carotid IMT (intima-medial thickness) at 240 degrees',
'Maximum carotid IMT (intima-medial thickness) at 240 degrees',
'Weight',
'Body fat percentage',
'Whole body fat mass',
'Whole body fat-free mass',
'Whole body water mass',
'Body mass index (BMI)',
'Basal metabolic rate',
'Impedance of whole body',
'Impedance of leg (right)',
'Impedance of leg (left)',
'Impedance of arm (right)',
'Impedance of arm (left)',
'Leg fat percentage (right)',
'Leg fat mass (right)',
'Leg fat-free mass (right)',
'Leg predicted mass (right)',
'Leg fat percentage (left)',
'Leg fat mass (left)',
'Leg fat-free mass (left)',
'Leg predicted mass (left)',
'Arm fat percentage (right)',
'Arm fat mass (right)',
'Arm fat-free mass (right)',
'Arm predicted mass (right)',
'Arm fat percentage (left)',
'Arm fat mass (left)',
'Arm fat-free mass (left)',
'Arm predicted mass (left)',
'Trunk fat percentage',
'Trunk fat mass',
'Trunk fat-free mass',
'Trunk predicted mass',
'Relative age of first facial hair',
'Age when periods started (menarche)',
'Birth weight of first child',
'Age at first live birth',
'Age at hysterectomy',
'Number of cigarettes previously smoked daily',
'White blood cell (leukocyte) count',
'Red blood cell (erythrocyte) count',
'Haemoglobin concentration',
'Haematocrit percentage',
'Mean corpuscular volume',
'Mean corpuscular haemoglobin',
'Red blood cell (erythrocyte) distribution width',
'Platelet count',
'Platelet crit',
'Mean platelet (thrombocyte) volume',
'Platelet distribution width',
'Lymphocyte count',
'Monocyte count',
'Neutrophill count',
'Eosinophill count',
'Lymphocyte percentage',
'Monocyte percentage',
'Neutrophill percentage',
'Eosinophill percentage',
'Reticulocyte percentage',
'Reticulocyte count',
'Mean reticulocyte volume',
'Mean sphered cell volume',
'Immature reticulocyte fraction',
'High light scatter reticulocyte percentage',
'High light scatter reticulocyte count',
'Forced vital capacity (FVC)',
'Forced expiratory volume in 1-second (FEV1)',
'Peak expiratory flow (PEF)',
'Ankle spacing width',
'Heel Broadband ultrasound attenuation, direct entry',
'Heel quantitative ultrasound index (QUI), direct entry',
'Heel bone mineral density (BMD)',
'Weight, manual entry',
'Age at menopause (last menstrual period)',
'Age asthma diagnosed',
'Age emphysema/chronic bronchitis diagnosed',
'Age deep-vein thrombosis (DVT, blood clot in leg) diagnosed',
'Diastolic blood pressure, automated reading',
'Systolic blood pressure, automated reading',
'Ankle spacing width (left)',
'Heel broadband ultrasound attenuation (left)',
'Heel quantitative ultrasound index (QUI), direct entry (left)',
'Heel bone mineral density (BMD) (left)',
'Heel bone mineral density (BMD) T-score, automated (left)',
'Ankle spacing width (right)',
'Heel broadband ultrasound attenuation (right)',
'Heel quantitative ultrasound index (QUI), direct entry (right)',
'Heel bone mineral density (BMD) (right)',
'Heel bone mineral density (BMD) T-score, automated (right)',
'Pulse rate',
'Round of numeric memory test',
'Maximum digits remembered correctly',
'Number of rounds of numeric memory test performed',
'Duration screen displayed',
'Hand grip strength (left)',
'Hand grip strength (right)',
'Waist circumference',
'Hip circumference',
'Standing height',
'Spherical power (right)',
'Spherical power (left)',
'3mm weak meridian (left)',
'6mm weak meridian (left)',
'6mm weak meridian (right)',
'3mm weak meridian (right)',
'Seated height',
'6mm asymmetry angle (left)',
'3mm strong meridian (right)',
'6mm strong meridian (right)',
'6mm strong meridian (left)',
'3mm strong meridian (left)',
'6mm asymmetry index (left)',
'6mm asymmetry index (right)',
'Intra-ocular pressure, corneal-compensated (right)',
'Intra-ocular pressure, Goldmann-correlated (right)',
'Corneal hysteresis (right)',
'Corneal resistance factor (right)',
'Intra-ocular pressure, corneal-compensated (left)',
'Intra-ocular pressure, Goldmann-correlated (left)',
'Corneal hysteresis (left)',
'Corneal resistance factor (left)',
'Heel bone mineral density (BMD) T-score, automated',
'Age completed full time education',
'Systolic blood pressure, manual reading',
'Diastolic blood pressure, manual reading',
'Pulse rate (during blood-pressure measurement)',
'Albumin/Globulin ratio',
'Diastolic blood pressure, automated reading, adjusted by medication',
'Diastolic blood pressure, combined automated + manual reading',
'Diastolic blood pressure, combined automated + manual reading, adjusted by medication',
'Diastolic blood pressure, manual reading, adjusted by medication',
'FEV1/FVC ratio',
'LDL direct, adjusted by medication',
'Mean arterial pressure, automated reading',
'Mean arterial pressure, automated reading, adjusted by medication',
'Mean arterial pressure, combined automated + manual reading',
'Mean arterial pressure, combined automated + manual reading, adjusted by medication',
'Mean arterial pressure, manual reading',
'Mean arterial pressure, manual reading, adjusted by medication',
'Non-albumin protein',
'Pulse pressure, automated reading',
'Pulse pressure, automated reading, adjusted by medication',
'Pulse pressure, combined automated + manual reading',
'Pulse pressure, combined automated + manual reading, adjusted by medication',
'Pulse pressure, manual reading',
'Pulse pressure, manual reading, adjusted by medication',
'Systolic blood pressure, automated reading, adjusted by medication',
'Systolic blood pressure, combined automated + manual reading',
'Systolic blood pressure, combined automated + manual reading, adjusted by medication',
'Systolic blood pressure, manual reading, adjusted by medication',
'Smoking status, ever vs never',
'Estimated glomerular filtration rate, serum creatinine',
'Estimated glomerular filtration rate, cystain C',
'Estimated glomerular filtration rate, serum creatinine + cystain C',
'pheno 48 / pheno 49']