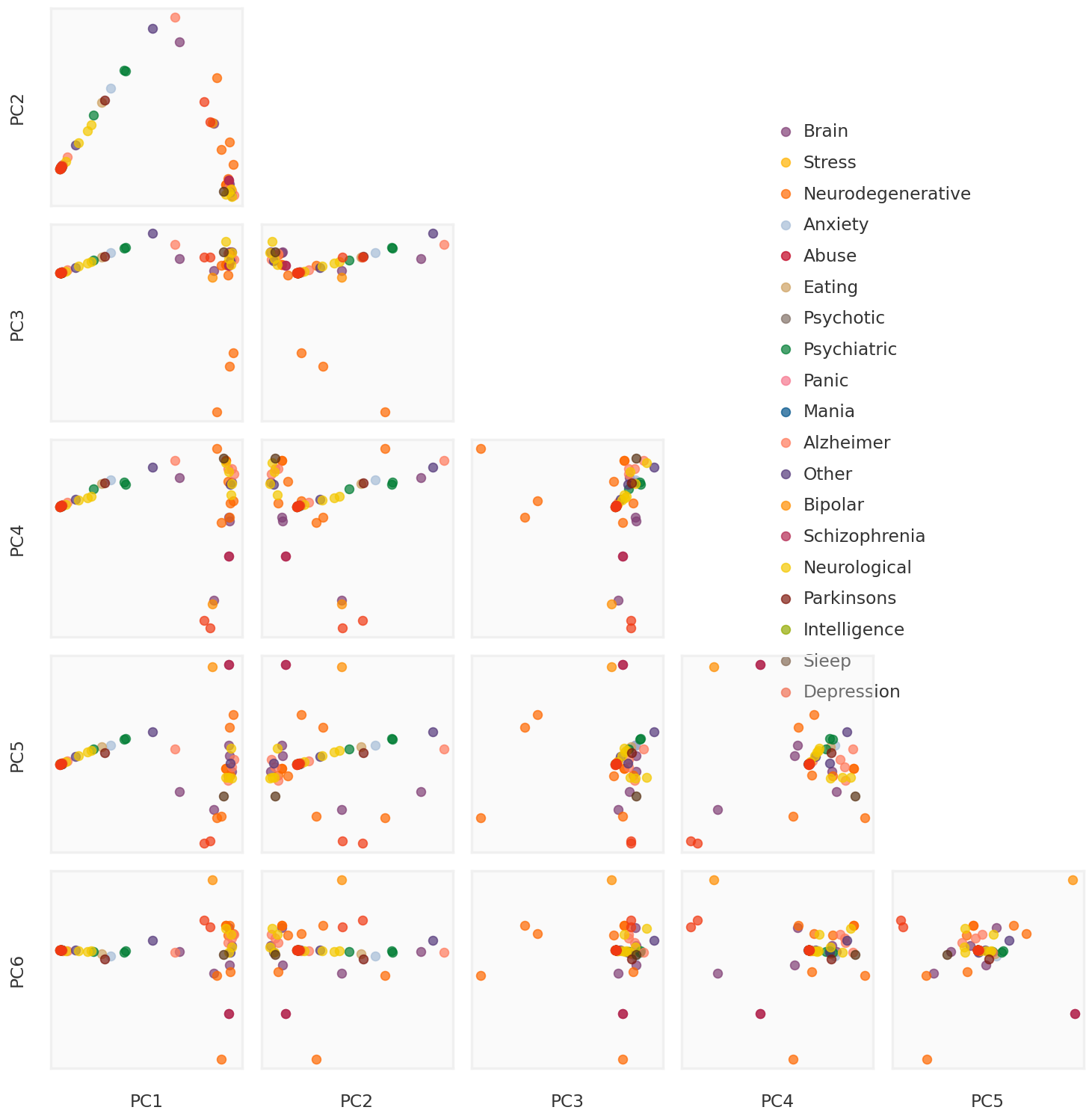
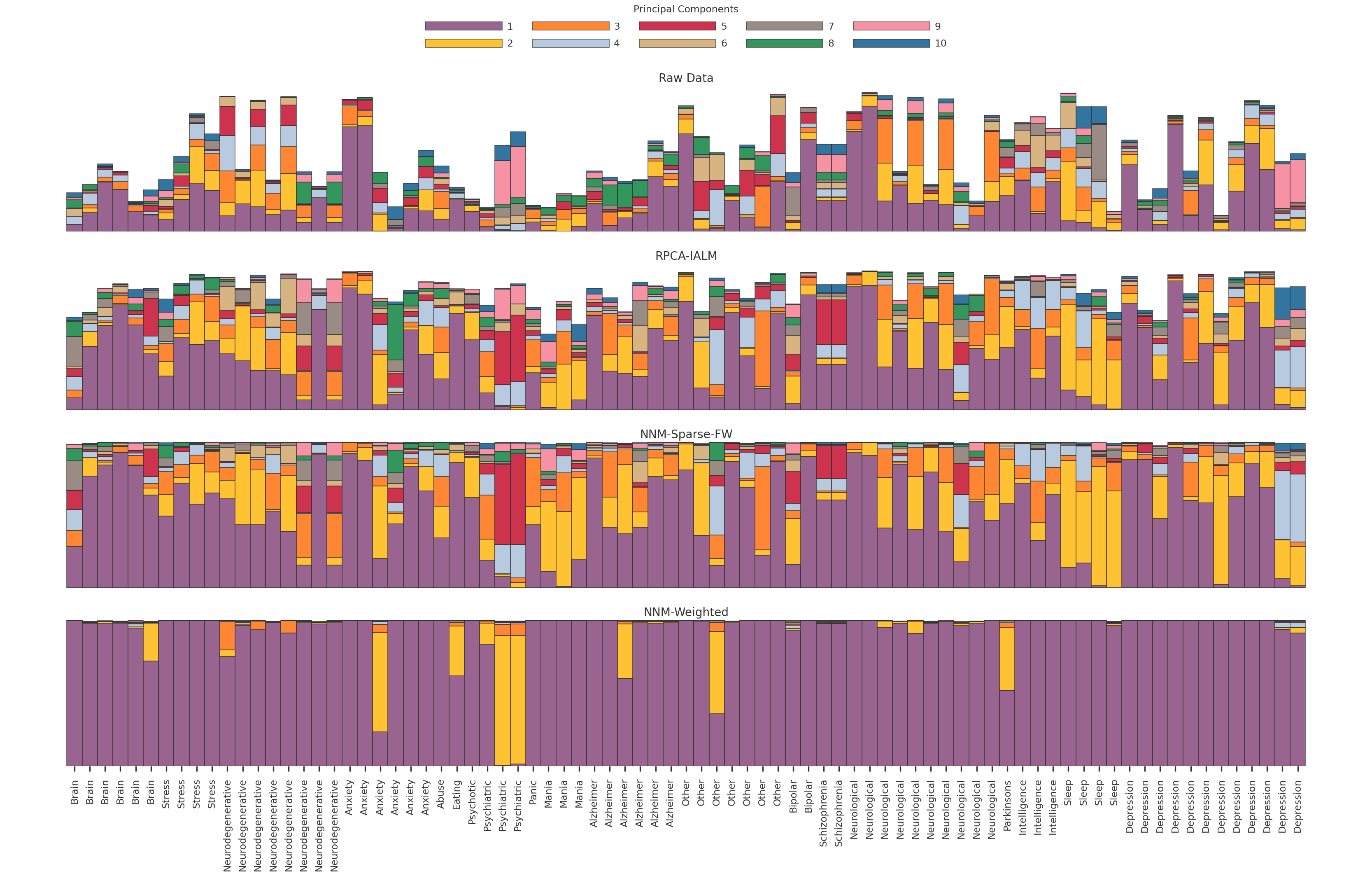
2023-11-27 14:23:56,518 | nnwmf.optimize.frankwolfe | INFO | Iteration 0. Step size 1.000. Duality Gap 3.45757e+06
2023-11-27 14:24:06,819 | nnwmf.optimize.frankwolfe | INFO | Iteration 100. Step size 0.006. Duality Gap 4026.97
2023-11-27 14:24:17,141 | nnwmf.optimize.frankwolfe | INFO | Iteration 200. Step size 0.002. Duality Gap 2322.95
2023-11-27 14:24:27,405 | nnwmf.optimize.frankwolfe | INFO | Iteration 300. Step size 0.002. Duality Gap 1759.15
2023-11-27 14:24:37,681 | nnwmf.optimize.frankwolfe | INFO | Iteration 400. Step size 0.003. Duality Gap 1873.36
2023-11-27 14:24:47,921 | nnwmf.optimize.frankwolfe | INFO | Iteration 500. Step size 0.001. Duality Gap 1094.16
2023-11-27 14:24:58,214 | nnwmf.optimize.frankwolfe | INFO | Iteration 600. Step size 0.001. Duality Gap 957.196
2023-11-27 14:25:08,429 | nnwmf.optimize.frankwolfe | INFO | Iteration 700. Step size 0.001. Duality Gap 925.217
2023-11-27 14:25:18,682 | nnwmf.optimize.frankwolfe | INFO | Iteration 800. Step size 0.001. Duality Gap 886.563
2023-11-27 14:25:28,948 | nnwmf.optimize.frankwolfe | INFO | Iteration 900. Step size 0.001. Duality Gap 718.781
2023-11-27 14:25:39,204 | nnwmf.optimize.frankwolfe | INFO | Iteration 1000. Step size 0.000. Duality Gap 354.201
2023-11-27 14:25:49,466 | nnwmf.optimize.frankwolfe | INFO | Iteration 1100. Step size 0.001. Duality Gap 647.135
2023-11-27 14:25:59,752 | nnwmf.optimize.frankwolfe | INFO | Iteration 1200. Step size 0.001. Duality Gap 556.836
2023-11-27 14:26:10,023 | nnwmf.optimize.frankwolfe | INFO | Iteration 1300. Step size 0.001. Duality Gap 742.643
2023-11-27 14:26:20,256 | nnwmf.optimize.frankwolfe | INFO | Iteration 1400. Step size 0.001. Duality Gap 576.098
2023-11-27 14:26:30,513 | nnwmf.optimize.frankwolfe | INFO | Iteration 1500. Step size 0.001. Duality Gap 439.903
2023-11-27 14:26:40,767 | nnwmf.optimize.frankwolfe | INFO | Iteration 1600. Step size 0.001. Duality Gap 508.349
2023-11-27 14:26:51,025 | nnwmf.optimize.frankwolfe | INFO | Iteration 1700. Step size 0.001. Duality Gap 496.27
2023-11-27 14:27:01,305 | nnwmf.optimize.frankwolfe | INFO | Iteration 1800. Step size 0.000. Duality Gap 375.604
2023-11-27 14:27:11,549 | nnwmf.optimize.frankwolfe | INFO | Iteration 1900. Step size 0.000. Duality Gap 474.234
2023-11-27 14:27:21,801 | nnwmf.optimize.frankwolfe | INFO | Iteration 2000. Step size 0.001. Duality Gap 455.82
2023-11-27 14:27:32,025 | nnwmf.optimize.frankwolfe | INFO | Iteration 2100. Step size 0.000. Duality Gap 376.674
2023-11-27 14:27:42,283 | nnwmf.optimize.frankwolfe | INFO | Iteration 2200. Step size 0.000. Duality Gap 343.212
2023-11-27 14:27:52,590 | nnwmf.optimize.frankwolfe | INFO | Iteration 2300. Step size 0.001. Duality Gap 436.296
2023-11-27 14:28:02,805 | nnwmf.optimize.frankwolfe | INFO | Iteration 2400. Step size 0.000. Duality Gap 358.472
2023-11-27 14:28:13,035 | nnwmf.optimize.frankwolfe | INFO | Iteration 2500. Step size 0.000. Duality Gap 222.27
2023-11-27 14:28:23,256 | nnwmf.optimize.frankwolfe | INFO | Iteration 2600. Step size 0.000. Duality Gap 275.813
2023-11-27 14:28:33,543 | nnwmf.optimize.frankwolfe | INFO | Iteration 2700. Step size 0.000. Duality Gap 257.902
2023-11-27 14:28:43,801 | nnwmf.optimize.frankwolfe | INFO | Iteration 2800. Step size 0.000. Duality Gap 250.31
2023-11-27 14:28:54,026 | nnwmf.optimize.frankwolfe | INFO | Iteration 2900. Step size 0.000. Duality Gap 213.307
2023-11-27 14:29:04,294 | nnwmf.optimize.frankwolfe | INFO | Iteration 3000. Step size 0.000. Duality Gap 315.758
2023-11-27 14:29:14,577 | nnwmf.optimize.frankwolfe | INFO | Iteration 3100. Step size 0.000. Duality Gap 212.123
2023-11-27 14:29:24,817 | nnwmf.optimize.frankwolfe | INFO | Iteration 3200. Step size 0.000. Duality Gap 120.102
2023-11-27 14:29:35,061 | nnwmf.optimize.frankwolfe | INFO | Iteration 3300. Step size 0.000. Duality Gap 295.67
2023-11-27 14:29:43,527 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:29:43,834 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:29:44,553 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:29:45,310 | nnwmf.optimize.frankwolfe | INFO | Iteration 3400. Step size 0.000. Duality Gap 193.651
2023-11-27 14:29:50,628 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:29:55,618 | nnwmf.optimize.frankwolfe | INFO | Iteration 3500. Step size 0.000. Duality Gap 287.296
2023-11-27 14:30:05,878 | nnwmf.optimize.frankwolfe | INFO | Iteration 3600. Step size 0.000. Duality Gap 33.0914
2023-11-27 14:30:16,112 | nnwmf.optimize.frankwolfe | INFO | Iteration 3700. Step size 0.000. Duality Gap 83.4953
2023-11-27 14:30:26,388 | nnwmf.optimize.frankwolfe | INFO | Iteration 3800. Step size 0.000. Duality Gap 204.393
2023-11-27 14:30:36,656 | nnwmf.optimize.frankwolfe | INFO | Iteration 3900. Step size 0.000. Duality Gap 157.054
2023-11-27 14:30:39,297 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:30:39,806 | nnwmf.optimize.frankwolfe | WARNING | Step Size is less than 0. Using last valid step size.
2023-11-27 14:30:46,890 | nnwmf.optimize.frankwolfe | INFO | Iteration 4000. Step size 0.000. Duality Gap 258.058
2023-11-27 14:30:57,170 | nnwmf.optimize.frankwolfe | INFO | Iteration 4100. Step size 0.000. Duality Gap 183.173
2023-11-27 14:31:07,378 | nnwmf.optimize.frankwolfe | INFO | Iteration 4200. Step size 0.000. Duality Gap 109.968