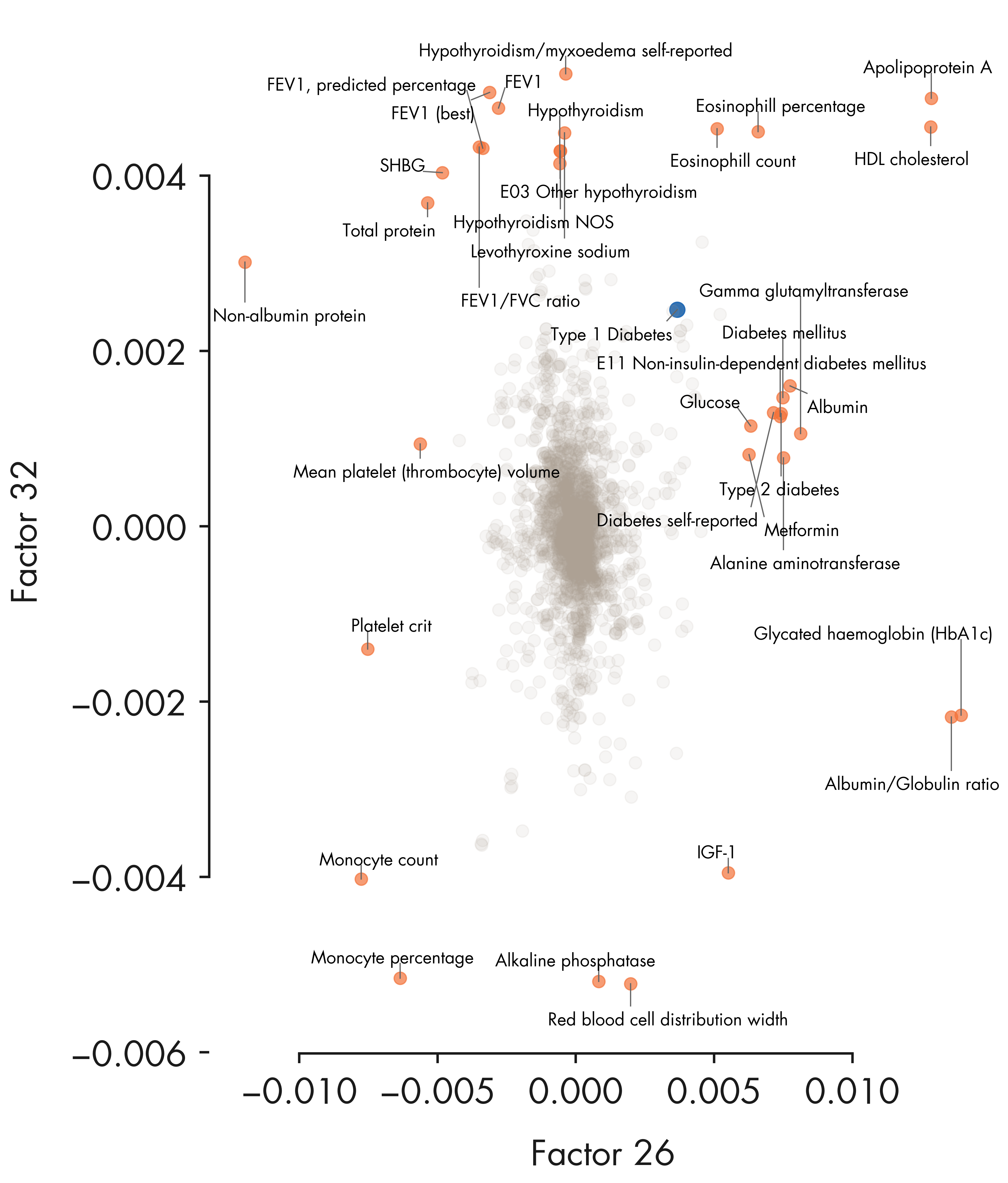
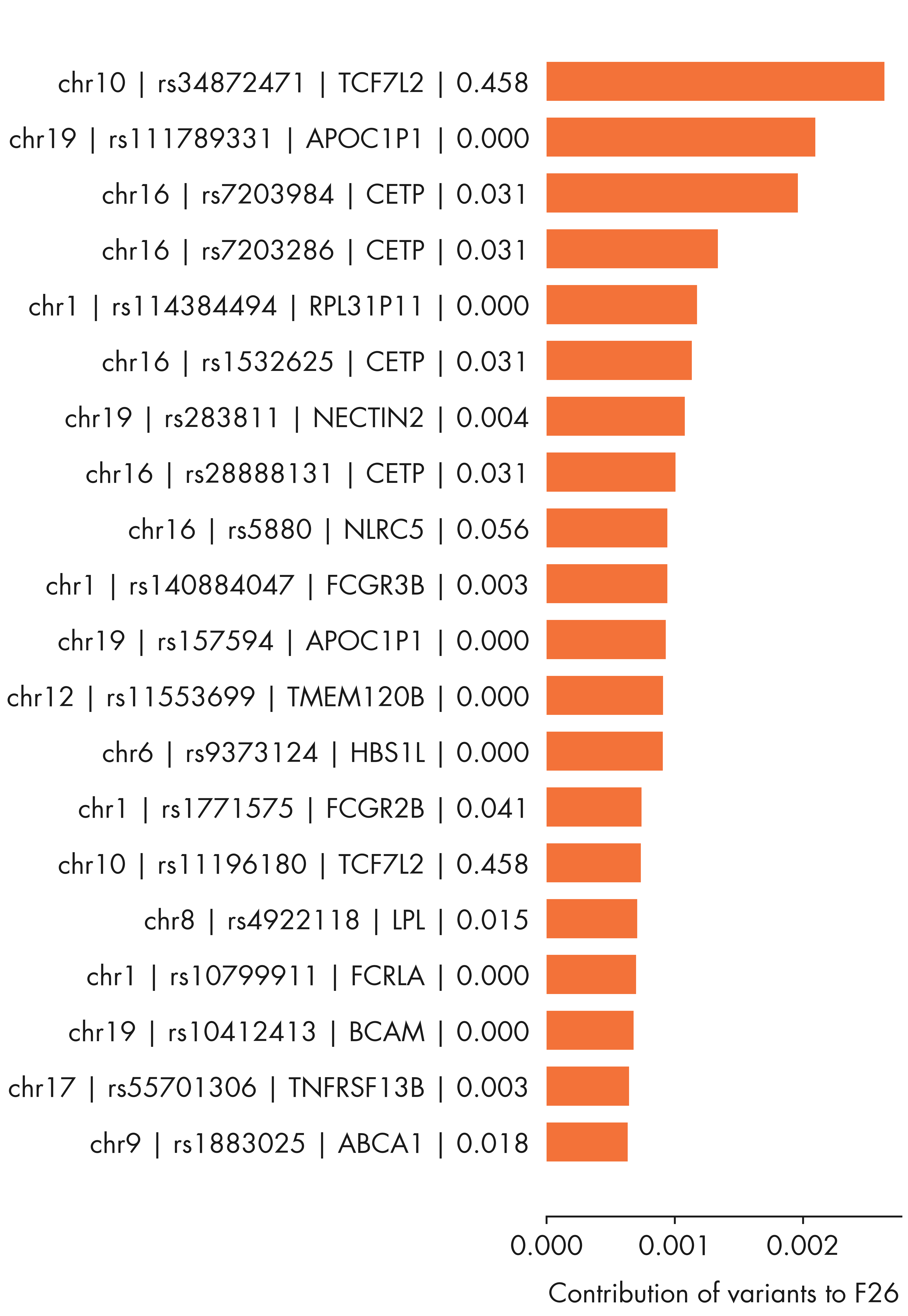
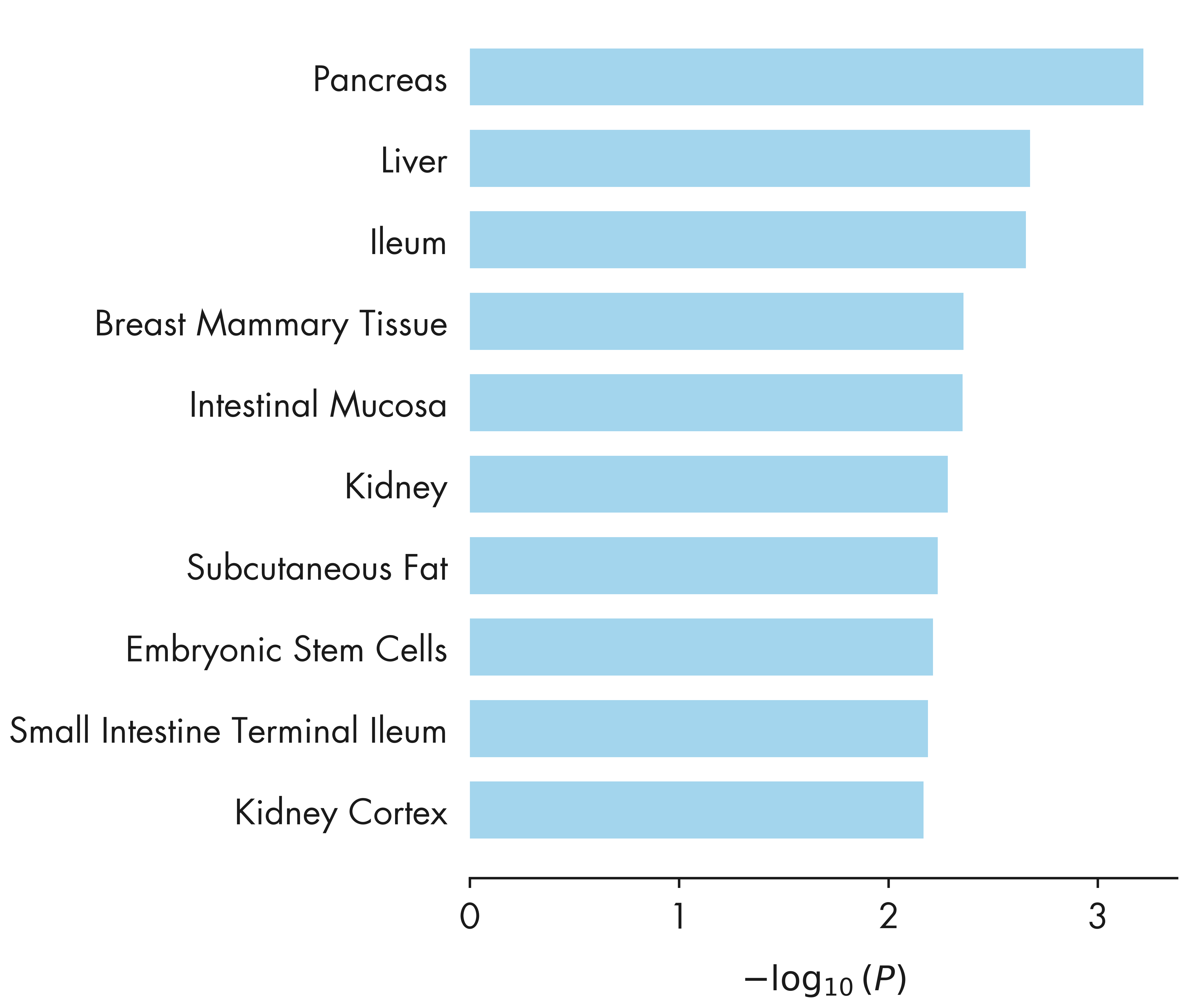
= plt.figure(figsize = (12 , 14 ))= fig.add_subplot(111 )# ax1.set_aspect('equal') = loadings[:, top_factor1]= - loadings[:, top_factor2]# Combine outliers in x-axis and y-axis # outlier_idx_x = np.where(iqr_outlier(xvals, axis = 0, bar = 5.0))[0] # outlier_idx_y = np.where(iqr_outlier(yvals, axis = 0, bar = 5.0))[0] = np.argsort(contribution_pheno[:, top_factor1])[::- 1 ][:20 ]= np.argsort(contribution_pheno[:, top_factor2])[::- 1 ][:20 ]= np.union1d(outlier_idx_x, outlier_idx_y)= np.mean(ax1.get_xlim())= np.setdiff1d(np.arange(xvals.shape[0 ]), outlier_idx)= ax1.scatter(xvals[outlier_idx], yvals[outlier_idx], alpha = 0.7 , s = 100 , color = nygc_colors['orange' ])= 150 , color = nygc_colors['blue' ])= 0.1 , s = 100 , color = nygc_colors['khaki' ])# # Mark using adjustText package # # https://github.com/Phlya/adjustText # annots = [] # for i in outlier_idx: # txt = trait_df.loc[trait_indices[i]]['description'].strip() # if 'intima-medial thickness' in txt: # continue # if xvals[i] > x_center: # annots += [ax1.annotate(txt, (xvals[i], yvals[i]), fontsize = 6, ha = 'right')] # else: # annots += [ax1.annotate(txt, (xvals[i], yvals[i]), fontsize = 6)] # # Adjust the annotations iteratively # adjust_text(annots, arrowprops=dict(arrowstyle='-', color = 'grey')) # Mark using textalloc package = []= []for i in outlier_idx:= trait_df_noRx.loc[trait_indices[i]]['short_description' ].strip()# Mark Type 1 diabetes "Type 1 Diabetes" )if len (text_idx_list) > 0 := np.array(text_idx_list)# x_scatter = xvals, y_scatter = yvals, = scatter_plot,= 14 , textcolor = 'black' , linecolor = nygc_colors['gray' ])- 0.010 , - 0.005 , 0.0 , 0.005 , 0.010 ])for side, border in ax1.spines.items():if side == 'left' :- 0.004 , 0.004 )elif side == 'bottom' :- 0.01 , 0.01 )else :False )f"Factor { top_factor1 + 1 } " )f"Factor { top_factor2 + 1 } " )'../plots/ashg24/phenotype_contributions_top2factors_t1d.png' , bbox_inches= 'tight' )