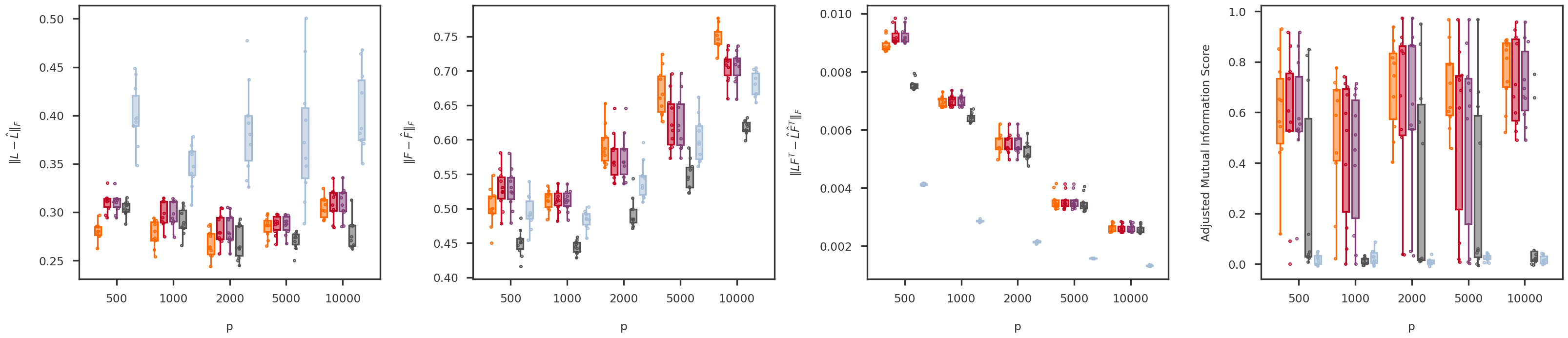
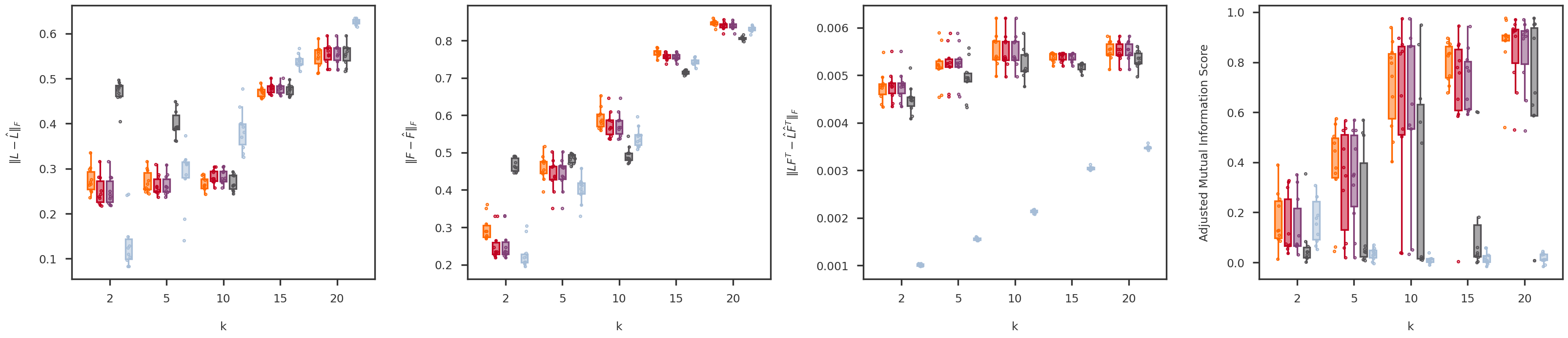
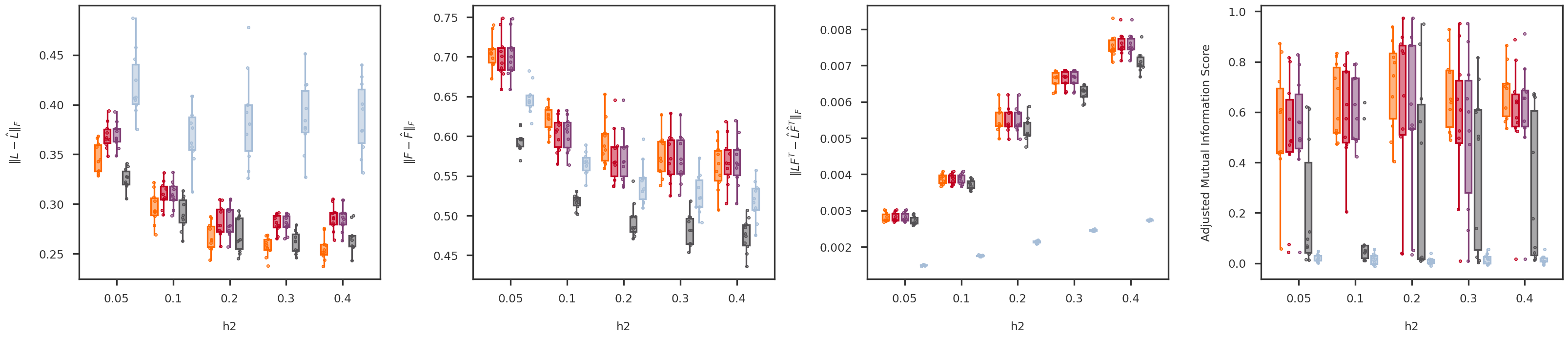
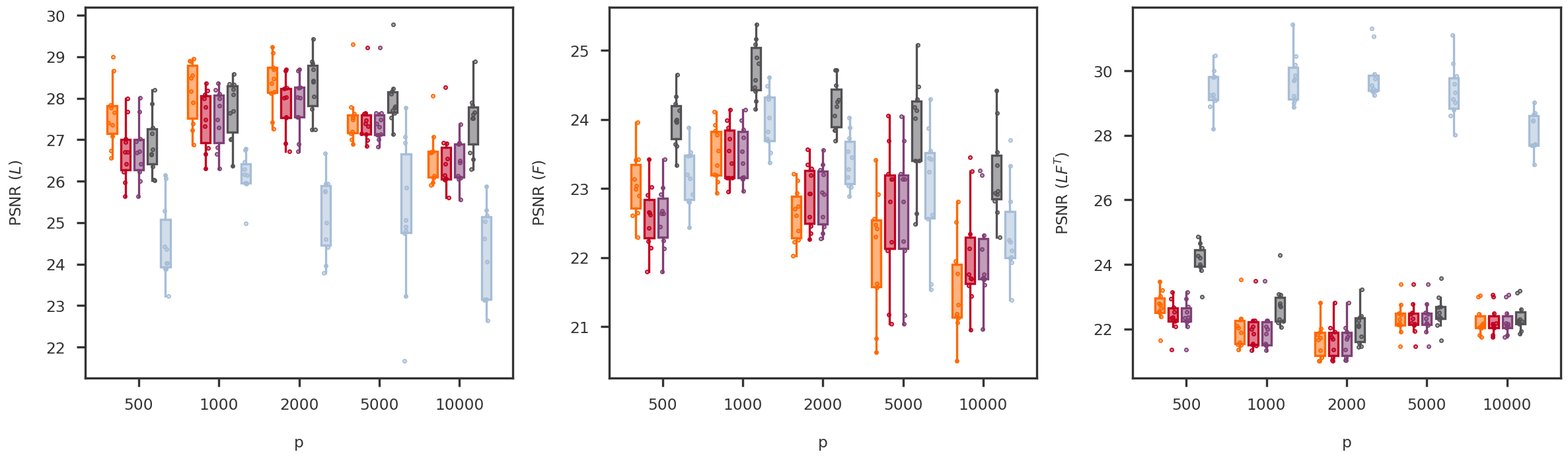
def random_jitter(xvals, yvals, d = 0.1 ):= [x + np.random.randn(len (y)) * d for x, y in zip (xvals, yvals)]return xjitterdef boxplot_scores(variable, variable_values, = methods, score_names = score_names,= dscout, method_colors = method_colors):= len (methods)= len (variable_values)= len (score_names)= 6 = (nscores * figh) + (nscores - 1 )= plt.figure(figsize = (figw, figh))= [fig.add_subplot(1 , nscores, x+ 1 ) for x in range (nscores)]= {x: None for x in methods.keys()}for i, (score_name, score_label) in enumerate (score_names.items()):= get_scores_from_dataframe(dscout, score_name, variable, variable_values)for j, mkey in enumerate (methods.keys()):= method_colors[mkey]= f'# { boxcolor[1 :]} 80' = dict (linewidth= 0 , color = boxcolor)= dict (linewidth= 2 , color = boxcolor)= dict (linewidth= 2 , color = boxcolor, facecolor = boxface)= dict (marker= 'o' , markerfacecolor= boxface, markersize= 3 , markeredgecolor = boxcolor)= [x * (nmethods + 1 ) + j for x in range (nvariables)]= axs[i].boxplot(scores[mkey], positions = xpos,= False , showfliers = False ,= 0.7 , patch_artist = True , notch = False ,= flierprops, boxprops = boxprops,= medianprops, whiskerprops = whiskerprops)= boxcolor, facecolor = boxface, linewidths = 1 , = 10 )= [x * (nmethods + 1 ) + (nmethods - 1 ) / 2 for x in range (nvariables)]= 0 - (nvariables - 1 ) / 2 #xlim_high = (nvariables - 1) * (nmethods + 1) + (nmethods - 1) + (nvariables - 1) / 2 = (nmethods + 1.5 ) * nvariables - 2.5 return axs, boxs= 'p' = [500 , 1000 , 2000 , 5000 , 10000 ]= boxplot_scores(variable, variable_values)