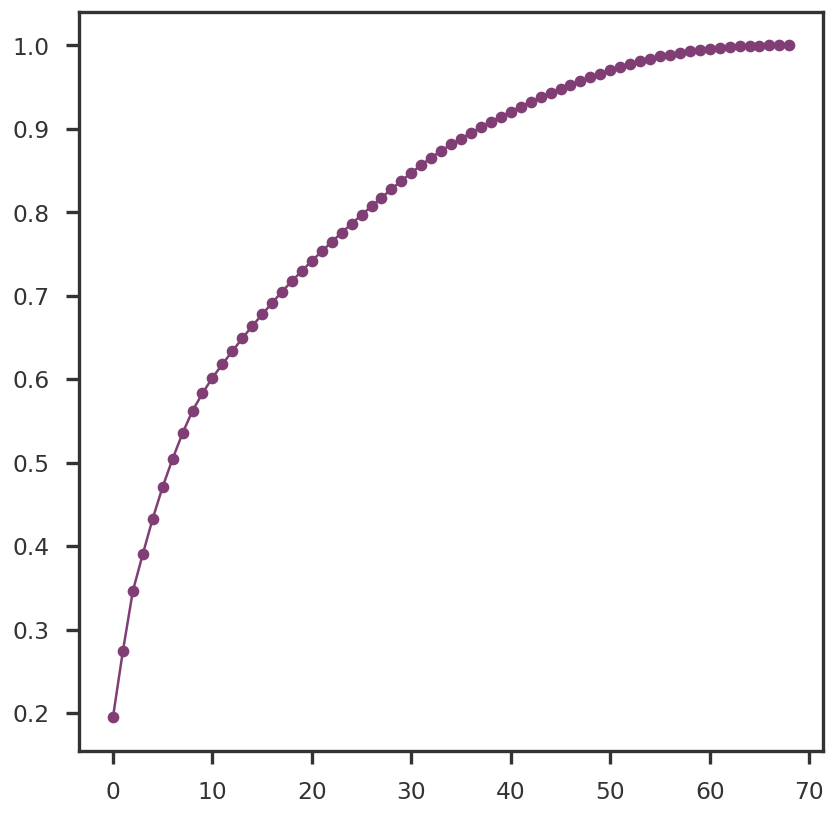
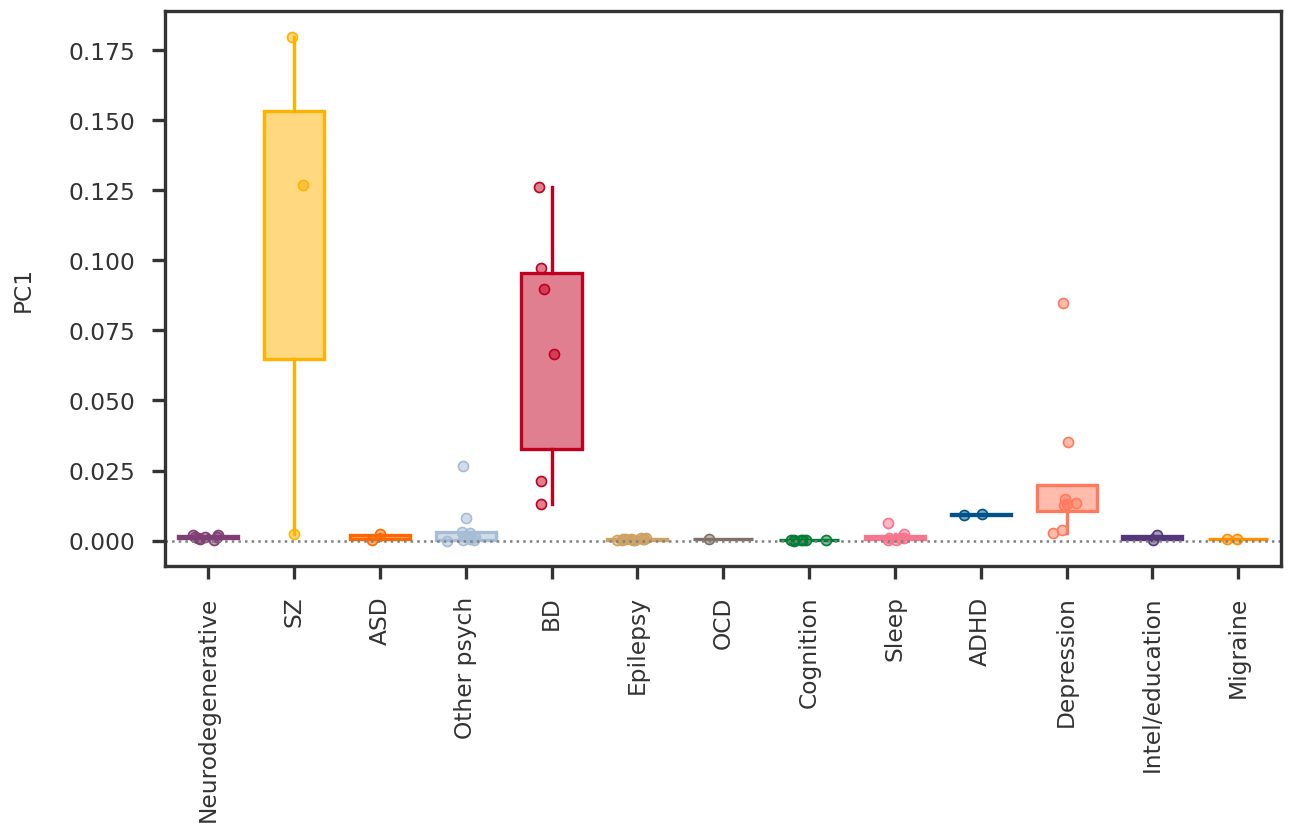
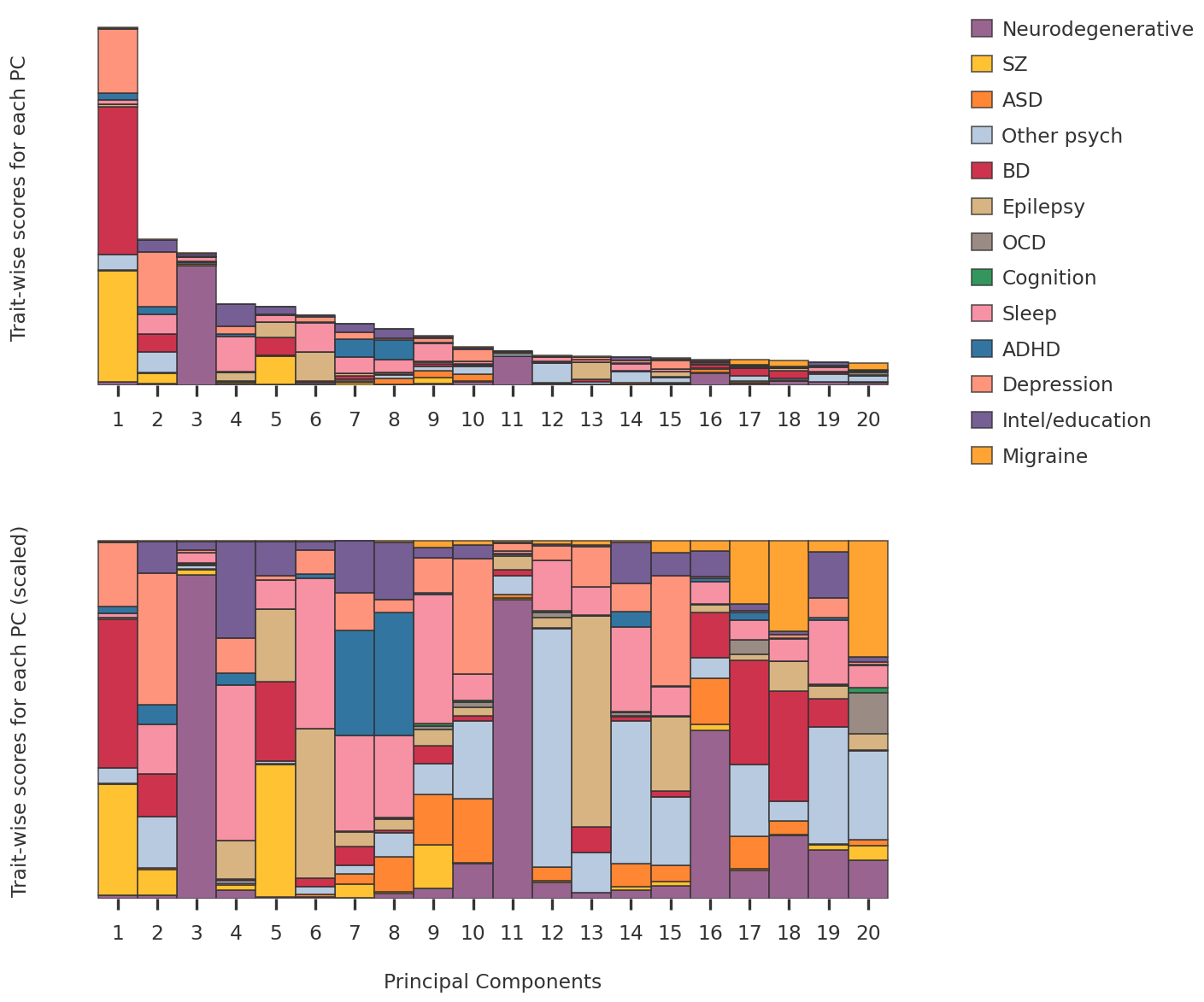
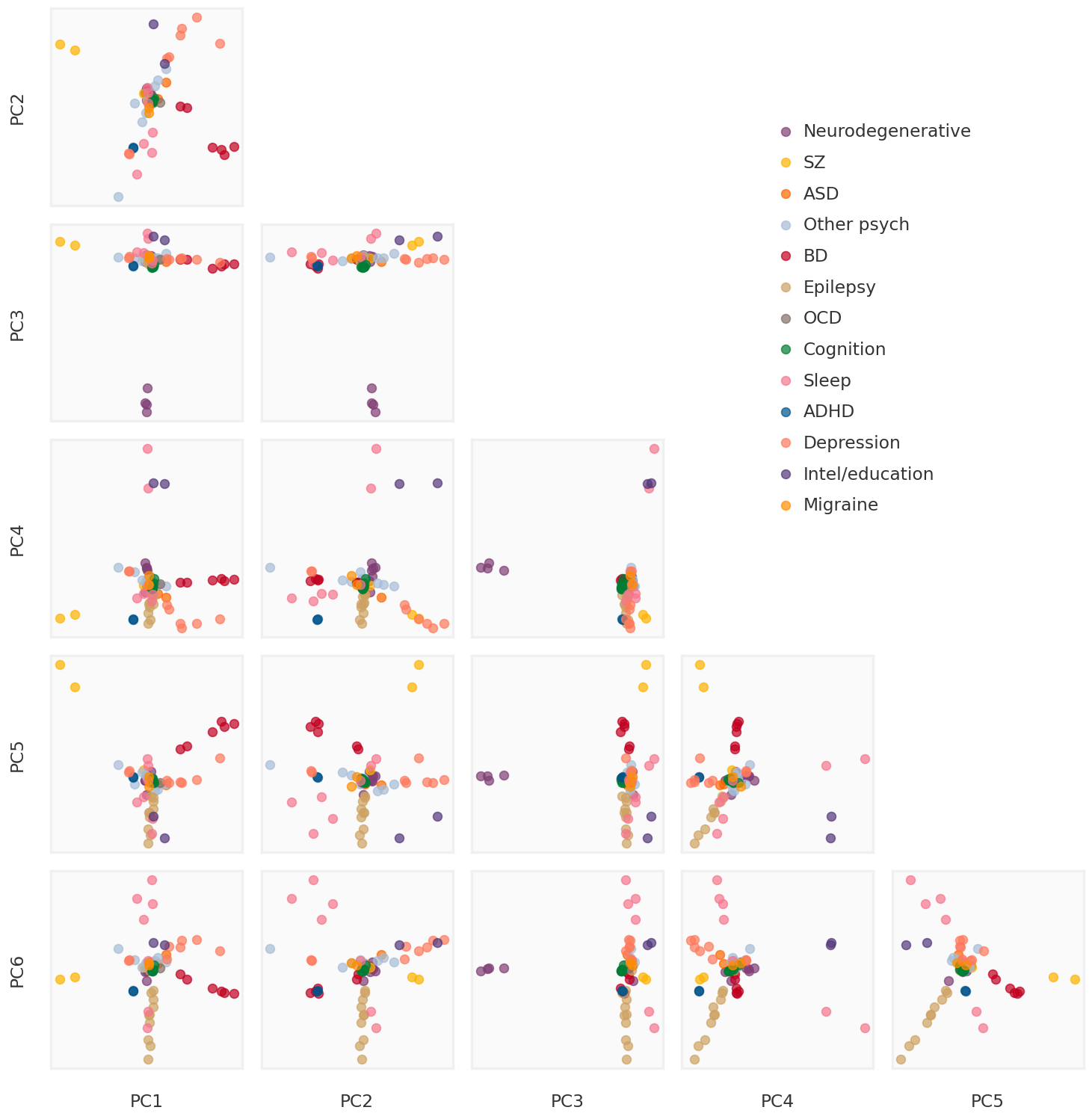
2023-08-08 23:54:07,309 | nnwmf.optimize.frankwolfe_cv | DEBUG | Cross-validation over 14 ranks.
2023-08-08 23:54:07,338 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 1 ...
2023-08-08 23:54:07,359 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1.0000
2023-08-08 23:54:09,099 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2.0000
2023-08-08 23:54:10,281 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4.0000
2023-08-08 23:54:14,327 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8.0000
2023-08-08 23:54:15,509 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 16.0000
2023-08-08 23:54:17,192 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 32.0000
2023-08-08 23:54:21,295 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 64.0000
2023-08-08 23:54:40,008 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 128.0000
2023-08-08 23:55:00,369 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 256.0000
2023-08-08 23:55:49,015 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 512.0000
2023-08-08 23:56:26,738 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1024.0000
2023-08-08 23:59:33,286 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2048.0000
2023-08-09 00:04:42,654 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4096.0000
2023-08-09 00:11:05,668 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8192.0000
2023-08-09 00:13:31,487 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 2 ...
2023-08-09 00:13:31,526 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1.0000
2023-08-09 00:13:32,692 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2.0000
2023-08-09 00:13:33,858 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4.0000
2023-08-09 00:13:35,041 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8.0000
2023-08-09 00:13:36,189 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 16.0000
2023-08-09 00:13:39,628 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 32.0000
2023-08-09 00:13:44,260 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 64.0000
2023-08-09 00:14:00,394 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 128.0000
2023-08-09 00:14:48,814 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 256.0000
2023-08-09 00:15:04,288 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 512.0000
2023-08-09 00:15:29,648 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1024.0000
2023-08-09 00:17:34,640 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2048.0000
2023-08-09 00:22:51,611 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4096.0000
2023-08-09 00:28:55,921 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8192.0000
2023-08-09 00:31:24,474 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 3 ...
2023-08-09 00:31:24,505 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1.0000
2023-08-09 00:31:25,651 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2.0000
2023-08-09 00:31:26,832 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4.0000
2023-08-09 00:31:28,004 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8.0000
2023-08-09 00:31:32,073 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 16.0000
2023-08-09 00:31:33,801 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 32.0000
2023-08-09 00:31:37,232 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 64.0000
2023-08-09 00:31:46,521 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 128.0000
2023-08-09 00:33:12,132 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 256.0000
2023-08-09 00:33:19,179 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 512.0000
2023-08-09 00:34:26,927 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1024.0000
2023-08-09 00:37:17,619 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2048.0000
2023-08-09 00:42:23,640 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4096.0000
2023-08-09 00:48:33,828 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8192.0000
2023-08-09 00:51:02,371 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 4 ...
2023-08-09 00:51:02,394 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1.0000
2023-08-09 00:51:03,577 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2.0000
2023-08-09 00:51:04,724 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4.0000
2023-08-09 00:51:06,453 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8.0000
2023-08-09 00:51:07,619 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 16.0000
2023-08-09 00:51:09,221 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 32.0000
2023-08-09 00:51:10,957 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 64.0000
2023-08-09 00:51:20,711 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 128.0000
2023-08-09 00:52:36,563 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 256.0000
2023-08-09 00:52:50,815 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 512.0000
2023-08-09 00:54:04,960 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1024.0000
2023-08-09 00:56:22,779 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2048.0000
2023-08-09 01:01:47,072 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4096.0000
2023-08-09 01:08:15,284 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8192.0000
2023-08-09 01:10:46,516 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 5 ...
2023-08-09 01:10:46,547 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1.0000
2023-08-09 01:10:47,748 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2.0000
2023-08-09 01:10:48,947 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4.0000
2023-08-09 01:10:50,135 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8.0000
2023-08-09 01:10:51,338 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 16.0000
2023-08-09 01:10:53,128 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 32.0000
2023-08-09 01:10:56,105 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 64.0000
2023-08-09 01:11:09,642 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 128.0000
2023-08-09 01:12:05,863 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 256.0000
2023-08-09 01:12:17,409 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 512.0000
2023-08-09 01:12:58,131 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 1024.0000
2023-08-09 01:15:24,342 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 2048.0000
2023-08-09 01:21:14,873 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 4096.0000
2023-08-09 01:27:29,020 | nnwmf.optimize.frankwolfe_cv | DEBUG | Rank 8192.0000