About
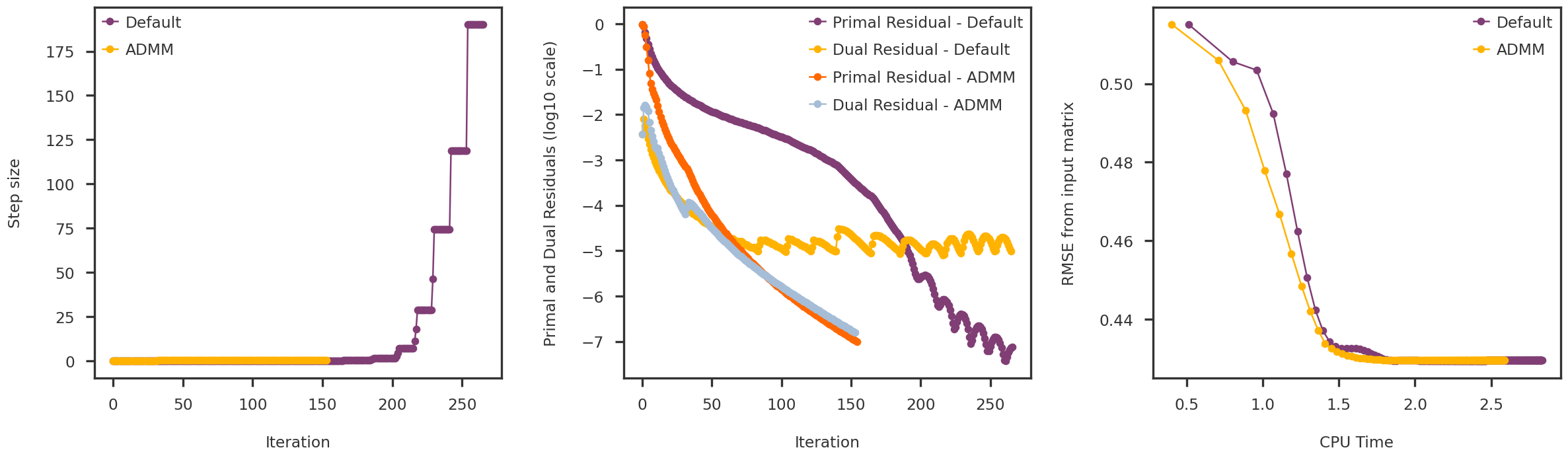
There are two alternate proposed methods for updating the penalty \mu on the residual. The penalty is equivalent to the step size. Here, we compare the two methods on a simulated dataset. The proposed methods are:
To-Do: Check stopping criterion of Boyd et. al. using Eq. 3.12.
Getting setup
Code
import numpy as npimport pandas as pdimport matplotlib.pyplot as pltfrom pymir import mpl_stylesheetfrom pymir import mpl_utils= 'black' , dpi = 120 , colors = 'kelly' )import sys"../utils/" )import histogram as mpy_histogramimport simulate as mpy_simulateimport plot_functions as mpy_plotfnfrom nnwmf.optimize import IALM
Simulate some toy data
= 4 # categories / class = 500 # N = 1000 # P = 40 # K
Code
= mpy_simulate.simulate(ngwas, nsnp, ntrait, nfctr, std = 0.5 , do_shift_mean = False )= mpy_simulate.do_standardize(Y, scale = False )= mpy_simulate.do_standardize(Y)= mpy_simulate.do_standardize(Y_true, scale = False )
Code
= list (range (len (sample_indices)))= [None for x in range (ngwas)]for k, idxs in enumerate (sample_indices):for i in idxs:= k
IALM with default update of step size
Code
= IALM(benchmark = True , max_iter = 1000 , show_progress = True )= Y_cent)
2023-07-28 13:24:39,445 | nnwmf.optimize.inexact_alm | INFO | Iteration 0. Primal residual 0.892391. Dual residual 0.00374201
2023-07-28 13:24:55,941 | nnwmf.optimize.inexact_alm | INFO | Iteration 100. Primal residual 0.0031319. Dual residual 1.12454e-05
2023-07-28 13:25:11,945 | nnwmf.optimize.inexact_alm | INFO | Iteration 200. Primal residual 2.69409e-06. Dual residual 1.09007e-05
IALM with ADMM update of step size
Code
= IALM(benchmark = True , max_iter = 1000 , mu_update_method= 'admm' , show_progress = True )= Y_cent)
2023-07-28 13:25:22,352 | nnwmf.optimize.inexact_alm | INFO | Iteration 0. Primal residual 0.892391. Dual residual 0.00374201
2023-07-28 13:25:38,124 | nnwmf.optimize.inexact_alm | INFO | Iteration 100. Primal residual 1.33026e-06. Dual residual 1.61284e-06
Results
Code
= plt.figure(figsize = (20 , 6 ))= fig.add_subplot(131 )= fig.add_subplot(132 )= fig.add_subplot(133 )range (len (rpca.mu_list_)), rpca.mu_list_, 'o-' , label = 'Default' )range (len (rpca_admm.mu_list_)), rpca_admm.mu_list_, 'o-' , label = 'ADMM' )"Iteration" )"Step size" )range (len (rpca.primal_res_)), np.log10(rpca.primal_res_), 'o-' , label = 'Primal Residual - Default' )range (len (rpca.dual_res_) - 1 ), np.log10(rpca.dual_res_[1 :]), 'o-' , label = 'Dual Residual - Default' )range (len (rpca_admm.primal_res_)), np.log10(rpca_admm.primal_res_), 'o-' , label = 'Primal Residual - ADMM' )range (len (rpca_admm.dual_res_) - 1 ), np.log10(rpca_admm.dual_res_[1 :]), 'o-' , label = 'Dual Residual - ADMM' )"Iteration" )"Primal and Dual Residuals (log10 scale)" )1 :])), rpca.rmse_[1 :], 'o-' , label = 'Default' )1 :])), rpca_admm.rmse_[1 :], 'o-' , label = 'ADMM' )"CPU Time" )"RMSE from input matrix" )= 2.0 )
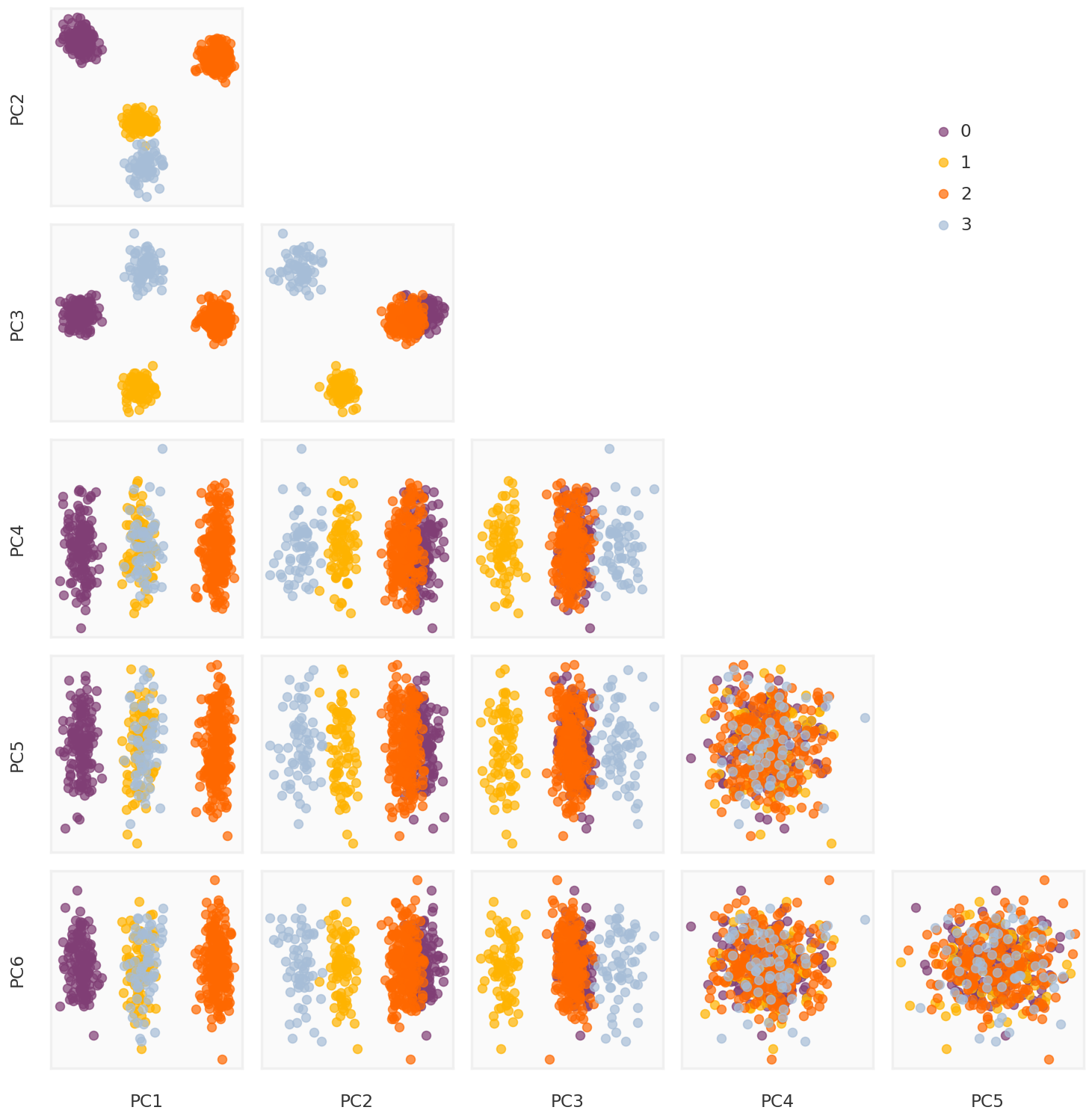
Principal Components for the low rank matrix
Default
Code
= mpy_simulate.do_standardize(rpca.L_, scale = False )= np.linalg.svd(Y_rpca_cent, full_matrices = False )= U_rpca @ np.diag(S_rpca)= mpy_plotfn.plot_principal_components(pcomps_rpca, class_labels, unique_labels)
ADMM
Code
= mpy_simulate.do_standardize(rpca_admm.L_, scale = False )= np.linalg.svd(Y_rpca_cent, full_matrices = False )= U_rpca @ np.diag(S_rpca)= mpy_plotfn.plot_principal_components(pcomps_rpca, class_labels, unique_labels)