About
Here, I check if the simulation benchmarking makes sense using simple examples. The idea is to run large scale simulations using pipelines. Before that, I want to look at simple examples.
Code
import numpy as npimport pandas as pdimport matplotlib.pyplot as pltfrom pymir import mpl_stylesheetfrom pymir import mpl_utils= 'black' , dpi = 120 , colors = 'kelly' )import sys"../utils/" )import histogram as mpy_histogramimport simulate as mpy_simulateimport plot_functions as mpy_plotfnfrom nnwmf.optimize import IALMfrom nnwmf.optimize import FrankWolfe_CVfrom nnwmf.optimize import FrankWolfe
= 4 # categories / class = 500 # N = 1000 # P = 40 # K
Code
= mpy_simulate.get_sample_indices(ntrait, ngwas, shuffle = False )= [x for _, x in sample_dict.items()]= [k for k, _ in sample_dict.items()]= [None for x in range (ngwas)]for k, x in sample_dict.items():for i in x:= k
Code
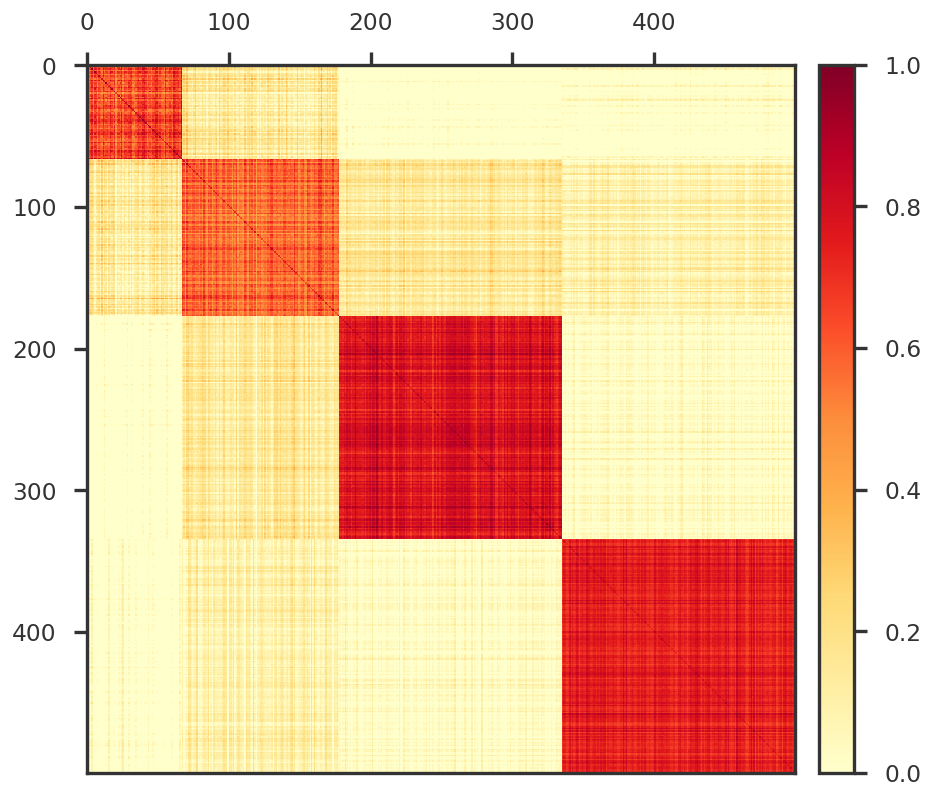
= mpy_simulate.simulate(ngwas, nsnp, ntrait, nfctr, sample_groups = sample_indices, std = 1.0 )= mpy_simulate.do_standardize(Y, scale = False )= plt.figure(figsize = (8 , 8 ))= fig.add_subplot(111 )
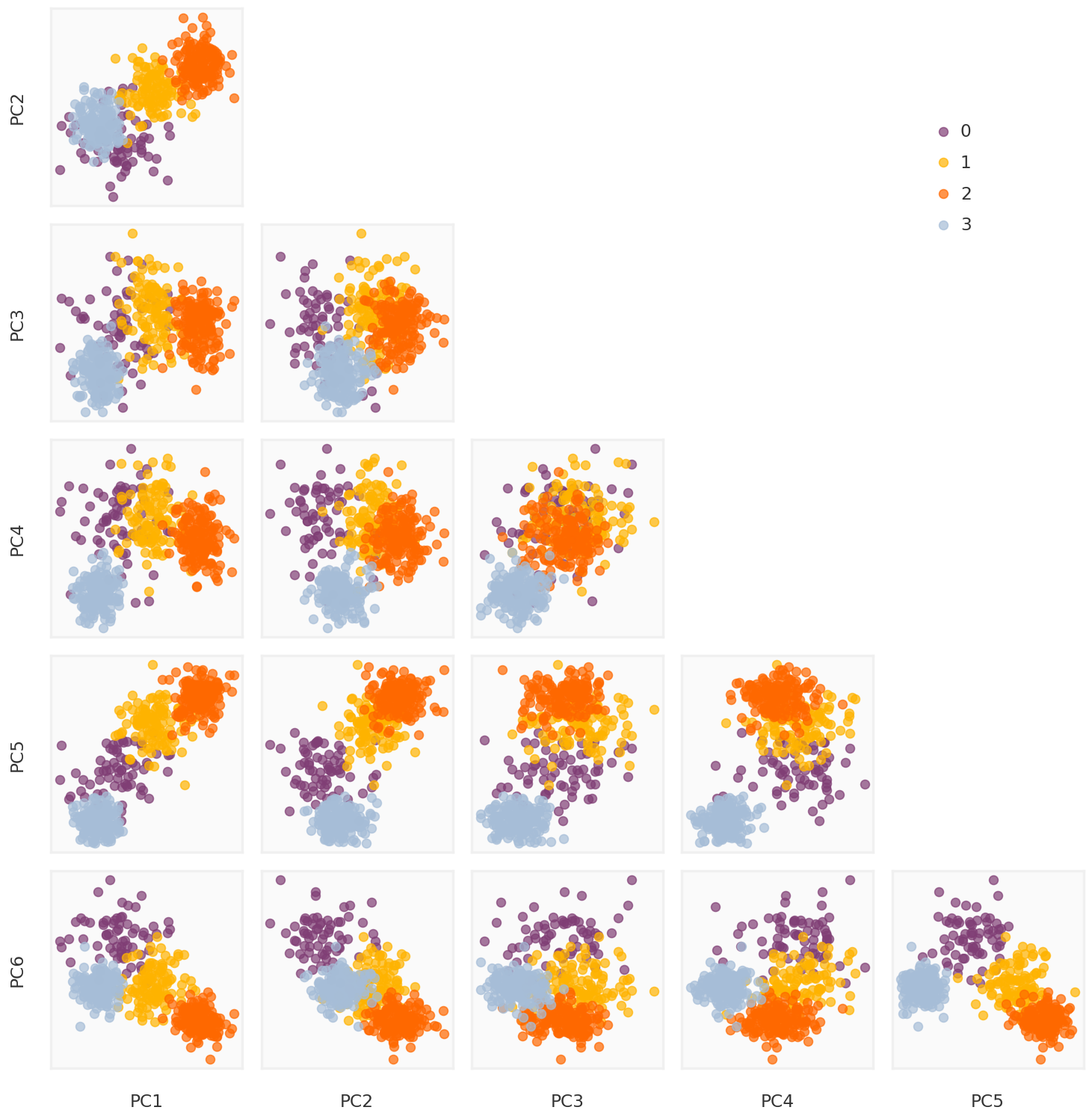
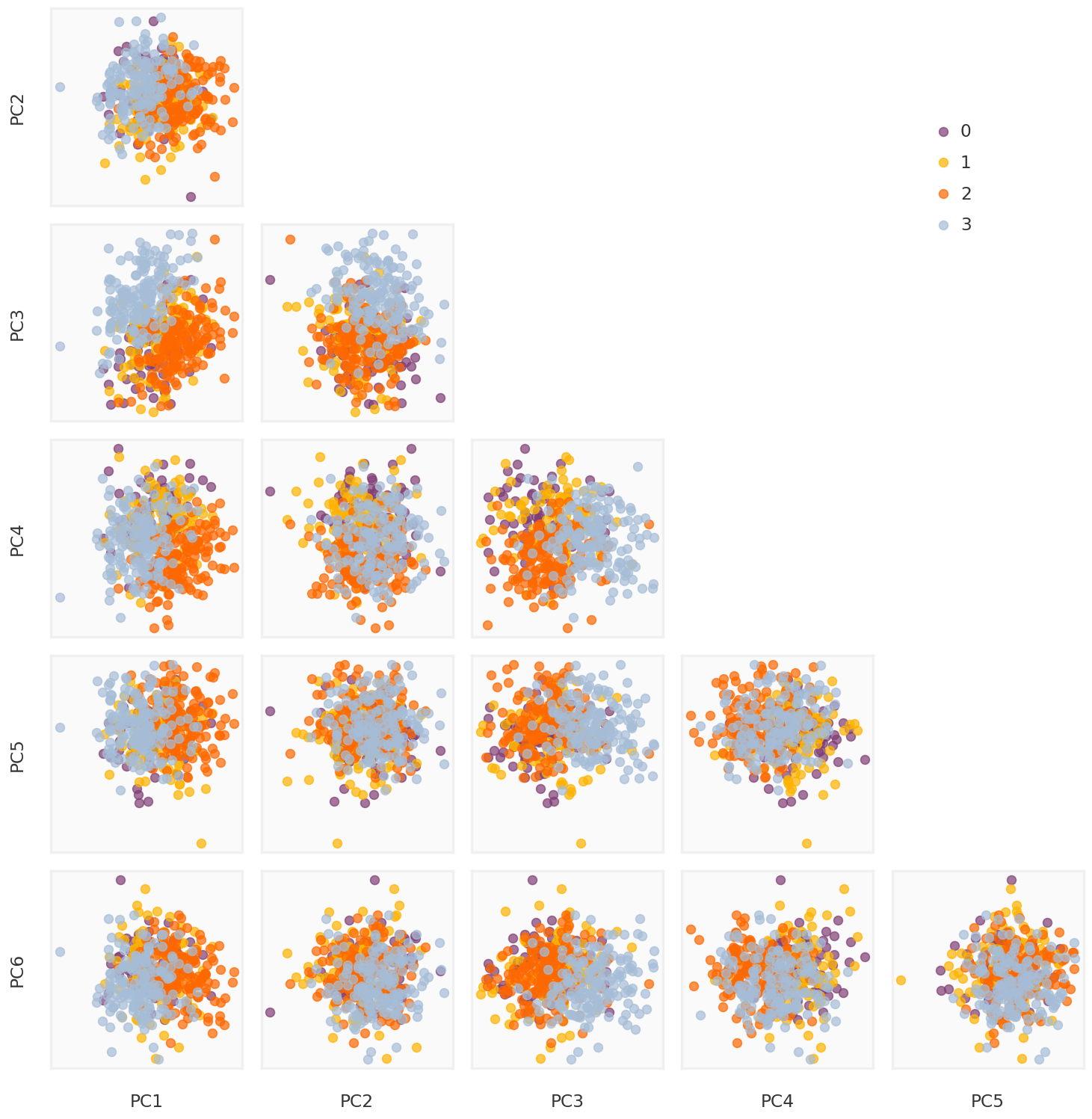
True components
Code
= mpy_plotfn.plot_principal_components(L, class_labels, unique_labels)
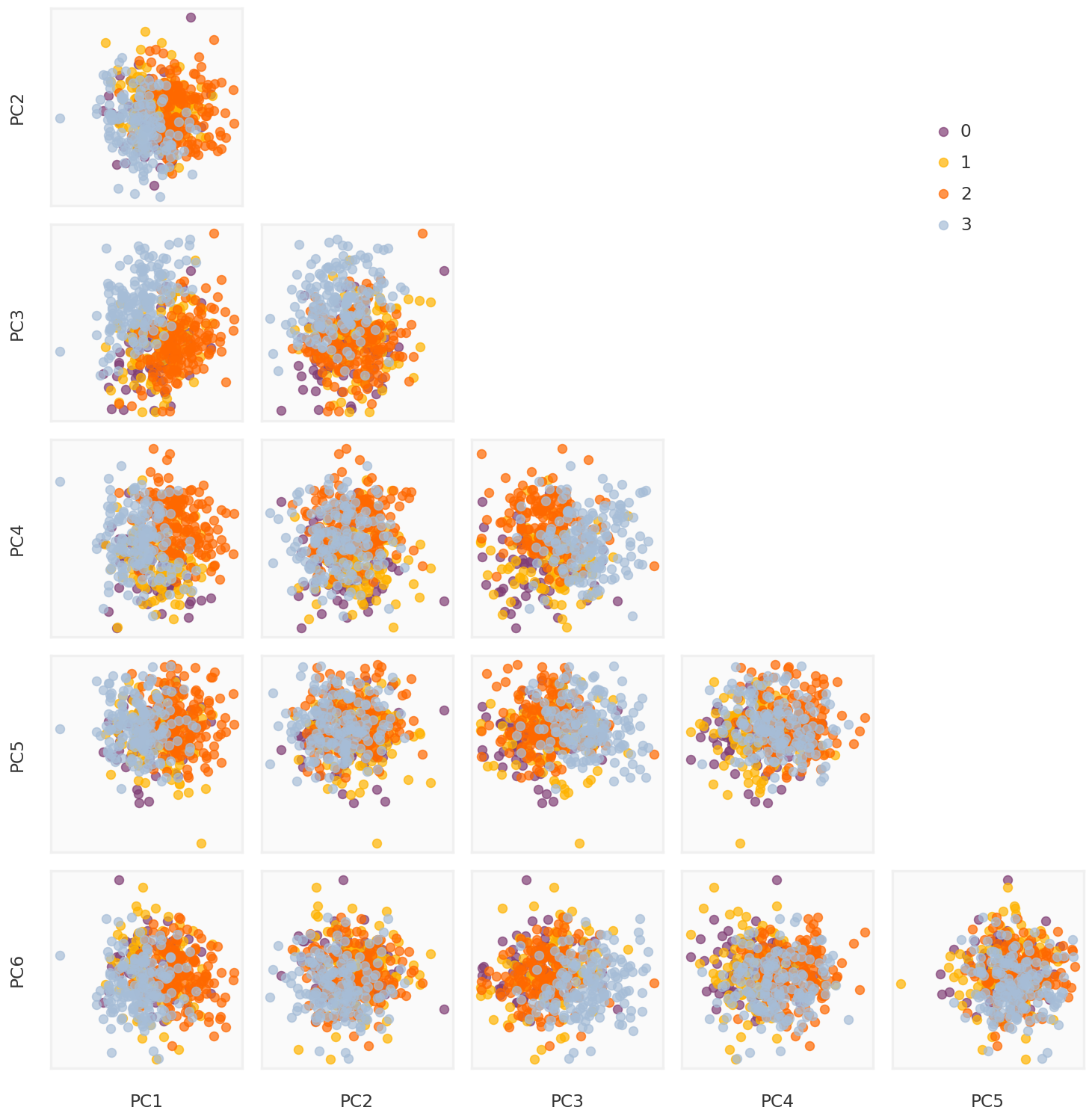
Truncated SVD
Code
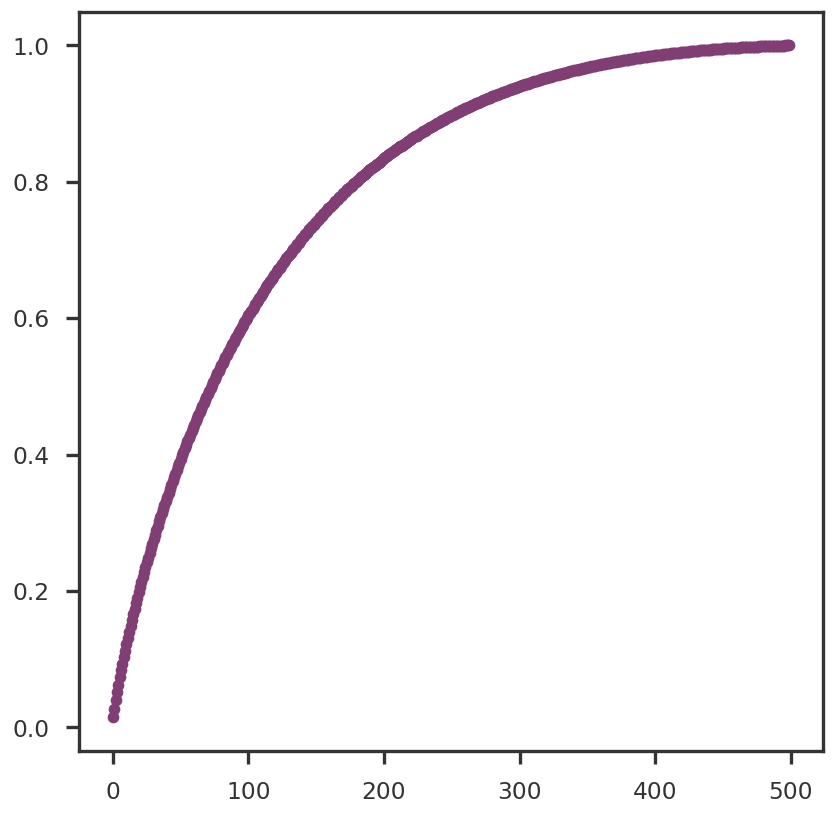
def truncated_SVD(X, thres = 0.9 ):= np.linalg.svd(X, full_matrices = False )= np.where(np.cumsum(S / np.sum (S)) >= thres)[0 ][0 ]= U[:, :k] @ np.diag(S[:k])return U, S, Vt, pcomps= truncated_SVD(Y_cent)= mpy_plotfn.plot_principal_components(pcomps_tsvd, class_labels, unique_labels)
Code
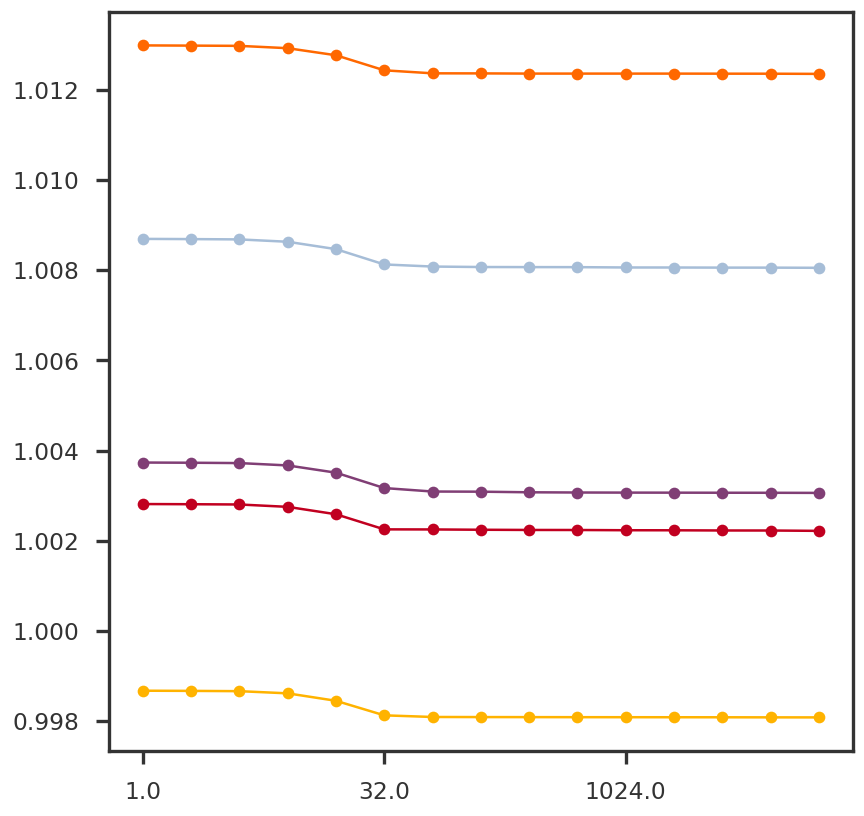
= np.square(S_tsvd)= plt.figure()= fig.add_subplot(111 )0 ]), np.cumsum(S2 / np.sum (S2)), 'o-' )
Nuclear Norm Minimization using Frank-Wolfe algorithm
Code
= FrankWolfe_CV(chain_init = True , reverse_path = False , kfolds = 2 )
Code
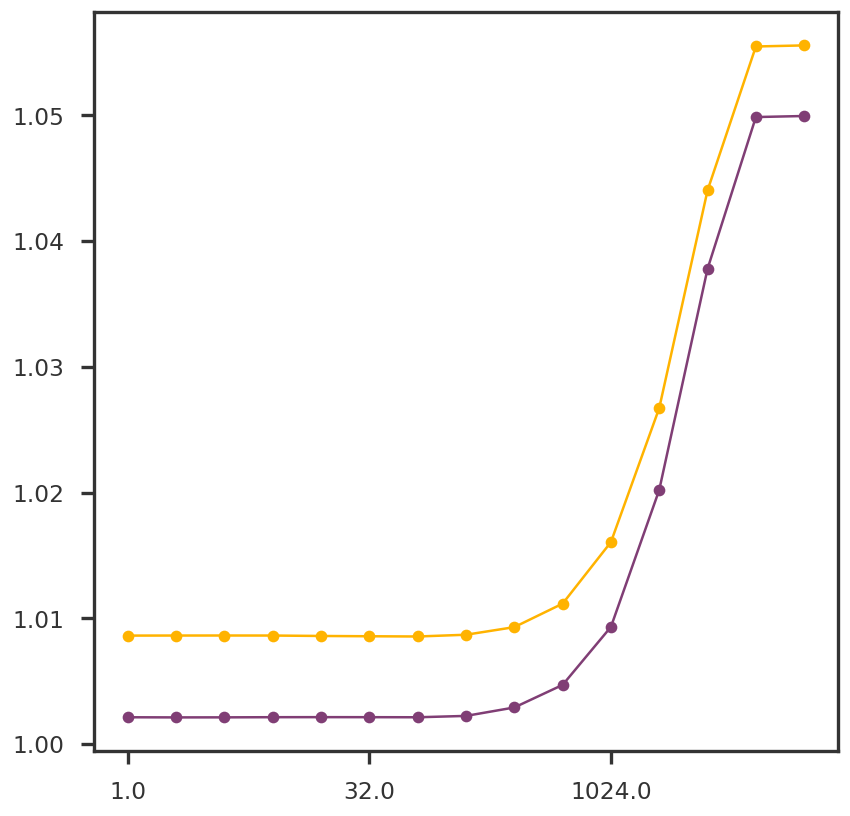
= plt.figure()= fig.add_subplot(111 )for k in range (2 ):#ax1.plot(np.log10(list(nnmcv.training_error.keys())), [x[k] for x in nnmcv.training_error.values()], 'o-') list (nnmcv.test_error.keys())), [x[k] for x in nnmcv.test_error.values()], 'o-' )= 'log10' , spacing = 'log2' )
Code
= 32.0 = FrankWolfe(show_progress = True , svd_max_iter = 50 , debug = True , suppress_warnings = True )= mpy_simulate.do_standardize(nnm.X, scale = False )= np.linalg.svd(Y_nnm_cent, full_matrices = False )= U_nnm @ np.diag(S_nnm)
2023-07-28 11:16:31,147 | nnwmf.optimize.frankwolfe | INFO | Iteration 0. Step size 1.000. Duality Gap 2742.83
Code
= mpy_plotfn.plot_principal_components(pcomps_nnm, class_labels, unique_labels)
Weighted Nuclear Norm Minimization
Code
= 1 / np.sqrt(noise_var)= np.column_stack([snp_weights for _ in range (ngwas)]).T= FrankWolfe_CV(chain_init = True , reverse_path = False , kfolds = 5 , debug = True )= weight)
2023-07-28 11:16:55,580 | nnwmf.optimize.frankwolfe_cv | DEBUG | Cross-validation over 15 ranks.
2023-07-28 11:16:55,604 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 1 ...
2023-07-28 11:17:13,426 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 2 ...
2023-07-28 11:17:35,075 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 3 ...
2023-07-28 11:17:57,035 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 4 ...
2023-07-28 11:18:16,387 | nnwmf.optimize.frankwolfe_cv | DEBUG | Fold 5 ...
Code
= plt.figure()= fig.add_subplot(111 )for k in range (5 ):#ax1.plot(np.log10(list(nnmcv.training_error.keys())), [x[k] for x in nnmcv.training_error.values()], 'o-') list (wnnmcv.test_error.keys())), [x[k] for x in wnnmcv.test_error.values()], 'o-' )= 'log10' , spacing = 'log2' )
Code
= 32.0 = FrankWolfe(show_progress = True , svd_max_iter = 50 , debug = True )= weight)= mpy_simulate.do_standardize(wnnm.X, scale = False )= np.linalg.svd(Y_wnnm_cent, full_matrices = False )= U_wnnm @ np.diag(S_wnnm)
2023-07-28 11:18:52,909 | nnwmf.optimize.frankwolfe | INFO | Iteration 0. Step size 0.154. Duality Gap 7.2392e+06
2023-07-28 11:18:55,519 | nnwmf.optimize.frankwolfe | INFO | Iteration 100. Step size 0.003. Duality Gap 100806
2023-07-28 11:18:58,015 | nnwmf.optimize.frankwolfe | INFO | Iteration 200. Step size 0.003. Duality Gap 42452.7
2023-07-28 11:19:00,500 | nnwmf.optimize.frankwolfe | INFO | Iteration 300. Step size 0.002. Duality Gap 31460.9
2023-07-28 11:19:02,997 | nnwmf.optimize.frankwolfe | INFO | Iteration 400. Step size 0.005. Duality Gap 20861.9
2023-07-28 11:19:05,500 | nnwmf.optimize.frankwolfe | INFO | Iteration 500. Step size 0.004. Duality Gap 16232.8
Code
= mpy_plotfn.plot_principal_components(pcomps_wnnm, class_labels, unique_labels)
Robust PCA
Code
= IALM()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[18], line 1
----> 1 rpca = IALM ( )
2 rpca. fit(Y_cent)
3 np. linalg. matrix_rank(rpca. L_)
File ~/work/nnwmf/src/nnwmf/optimize/inexact_alm.py:85 , in IALM.__init__ (self, rho, tau, mu_update_method, max_iter, primal_tol, dual_tol)
82 assert rho > 1
83 assert tau > 1
---> 85 self . lmb_ = lmb
86 self . rho_ = rho
87 self . max_iter_ = max_iter
NameError : name 'lmb' is not defined
Code
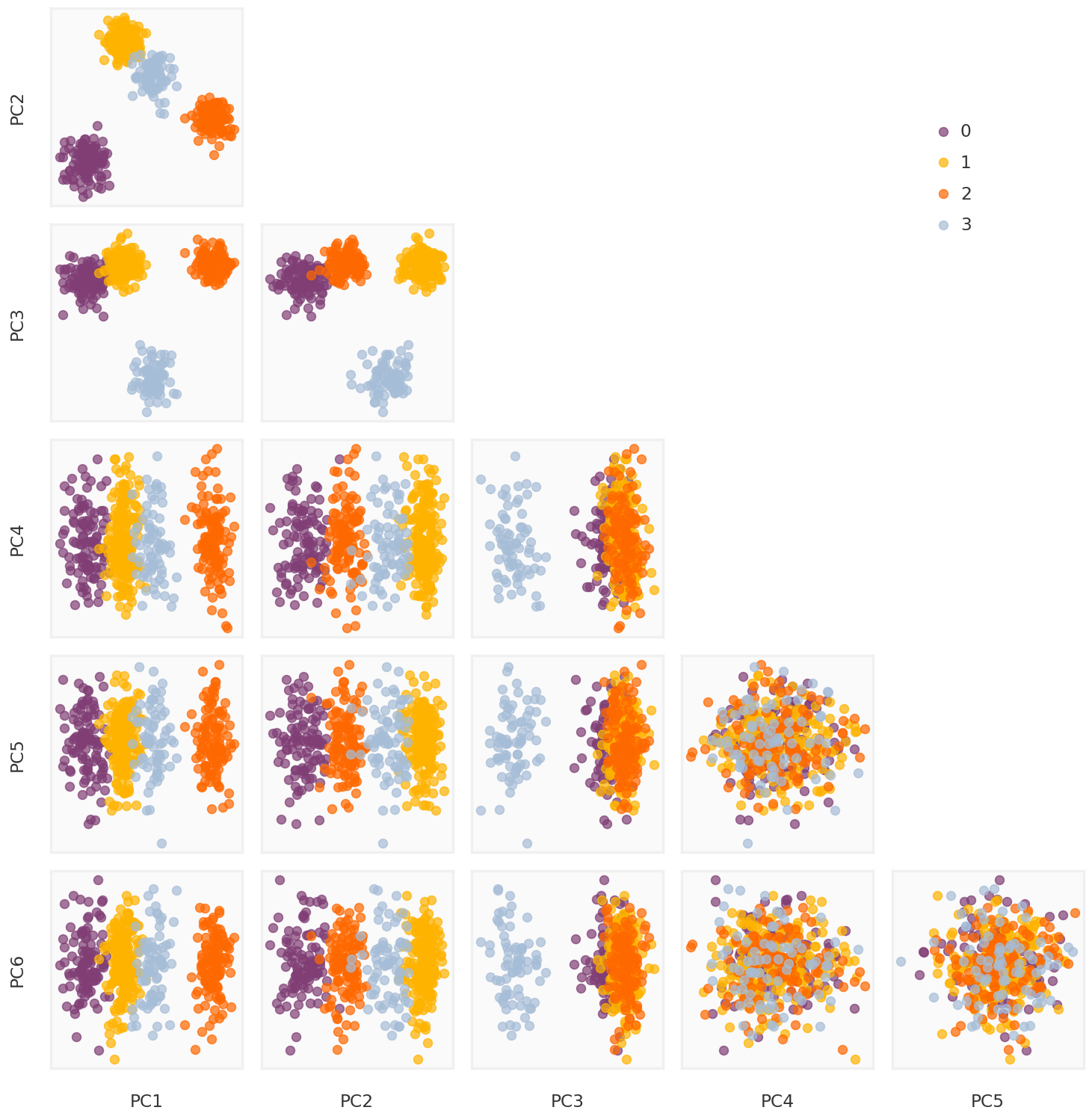
= mpy_simulate.do_standardize(L_rpca, scale = False )= np.linalg.svd(Y_rpca_cent, full_matrices = False )= U_rpca @ np.diag(S_rpca)= mpy_plotfn.plot_principal_components(pcomps_rpca, class_labels, unique_labels)
Code
def get_rmse(original, recovered, mask = None ):if mask is None := np.ones(original.shape)= np.sum (mask)= np.sum (np.square((original - recovered) * mask)) / nreturn np.sqrt(mse)= mpy_simulate.do_standardize(Y_true, scale = False )= mpy_simulate.do_standardize(L_rpca, scale = False )= get_rmse(Y_true_cent, Y_nnm_cent)= get_rmse(Y_true_cent, Y_wnnm_cent)= get_rmse(Y_true_cent, Y_rpca_cent)print (f" { rmse_nnm:.4f} \t Nuclear Norm Minimization" )print (f" { rmse_wnnm:.4f} \t Weighted Nuclear Norm Minimization" )print (f" { rmse_rpca:.4f} \t Robust PCA" )
0.1448 Nuclear Norm Minimization
0.1427 Weighted Nuclear Norm Minimization
0.1054 Robust PCA
Code
def get_psnr(original, recovered):= original.shape= np.square(np.max (original) - np.min (original))= np.sum (np.square(recovered - original)) / (n * p)= 10 * np.log10(maxsig2 / mse)return res= get_psnr(Y_true_cent, Y_nnm_cent)= get_psnr(Y_true_cent, Y_wnnm_cent)= get_psnr(Y_true_cent, Y_rpca_cent)print (f" { psnr_nnm:.4f} \t Nuclear Norm Minimization" )print (f" { psnr_wnnm:.4f} \t Weighted Nuclear Norm Minimization" )print (f" { psnr_rpca:.4f} \t Robust PCA" )
21.2372 Nuclear Norm Minimization
21.3642 Weighted Nuclear Norm Minimization
23.9949 Robust PCA
Code
from sklearn.cluster import AgglomerativeClusteringfrom sklearn import metrics as skmetricsdef get_adjusted_MI_score(pcomp, class_labels):= skmetrics.pairwise.pairwise_distances(pcomp, metric= 'euclidean' )= AgglomerativeClustering(n_clusters = 4 , linkage = 'average' , metric = 'precomputed' )= model.fit_predict(distance_matrix)return skmetrics.adjusted_mutual_info_score(class_labels, class_pred)= get_adjusted_MI_score(pcomps_nnm, class_labels)= get_adjusted_MI_score(pcomps_wnnm, class_labels)= get_adjusted_MI_score(pcomps_rpca, class_labels)print (f" { adjusted_mi_nnm:.4f} \t Nuclear Norm Minimization" )print (f" { adjusted_mi_wnnm:.4f} \t Weighted Nuclear Norm Minimization" )print (f" { adjusted_mi_rpca:.4f} \t Robust PCA" )
-0.0006 Nuclear Norm Minimization
1.0000 Weighted Nuclear Norm Minimization
1.0000 Robust PCA