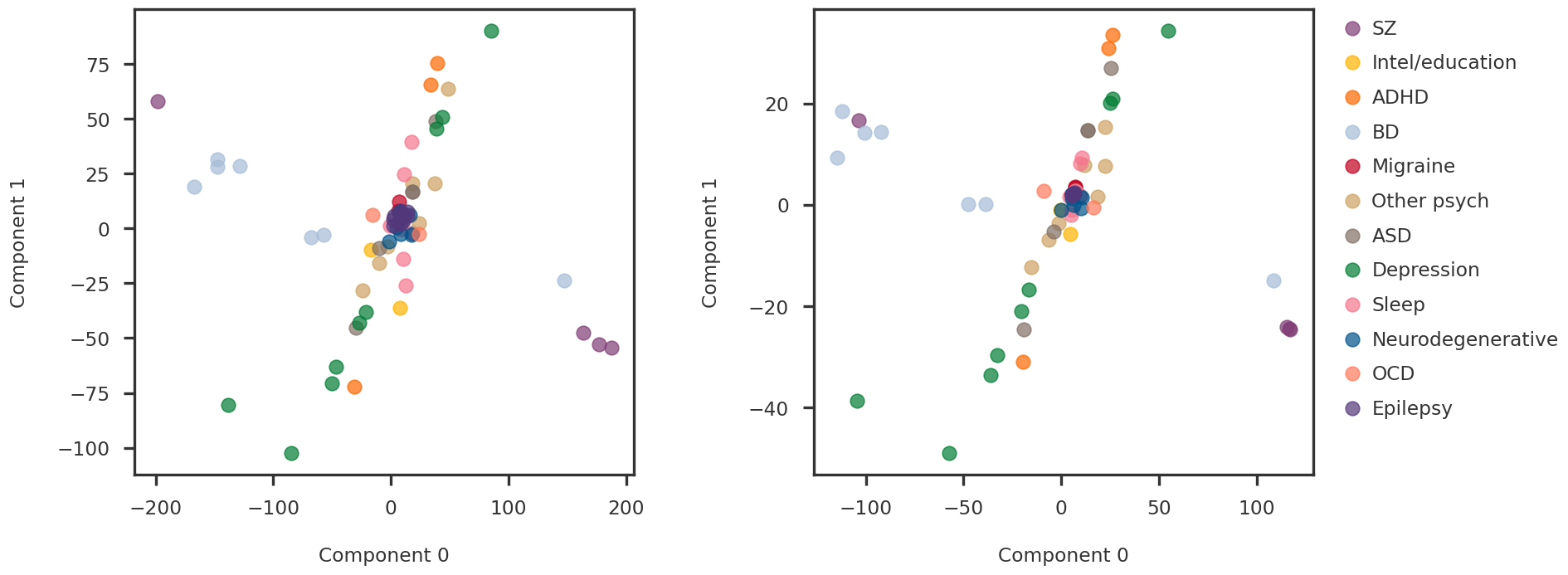
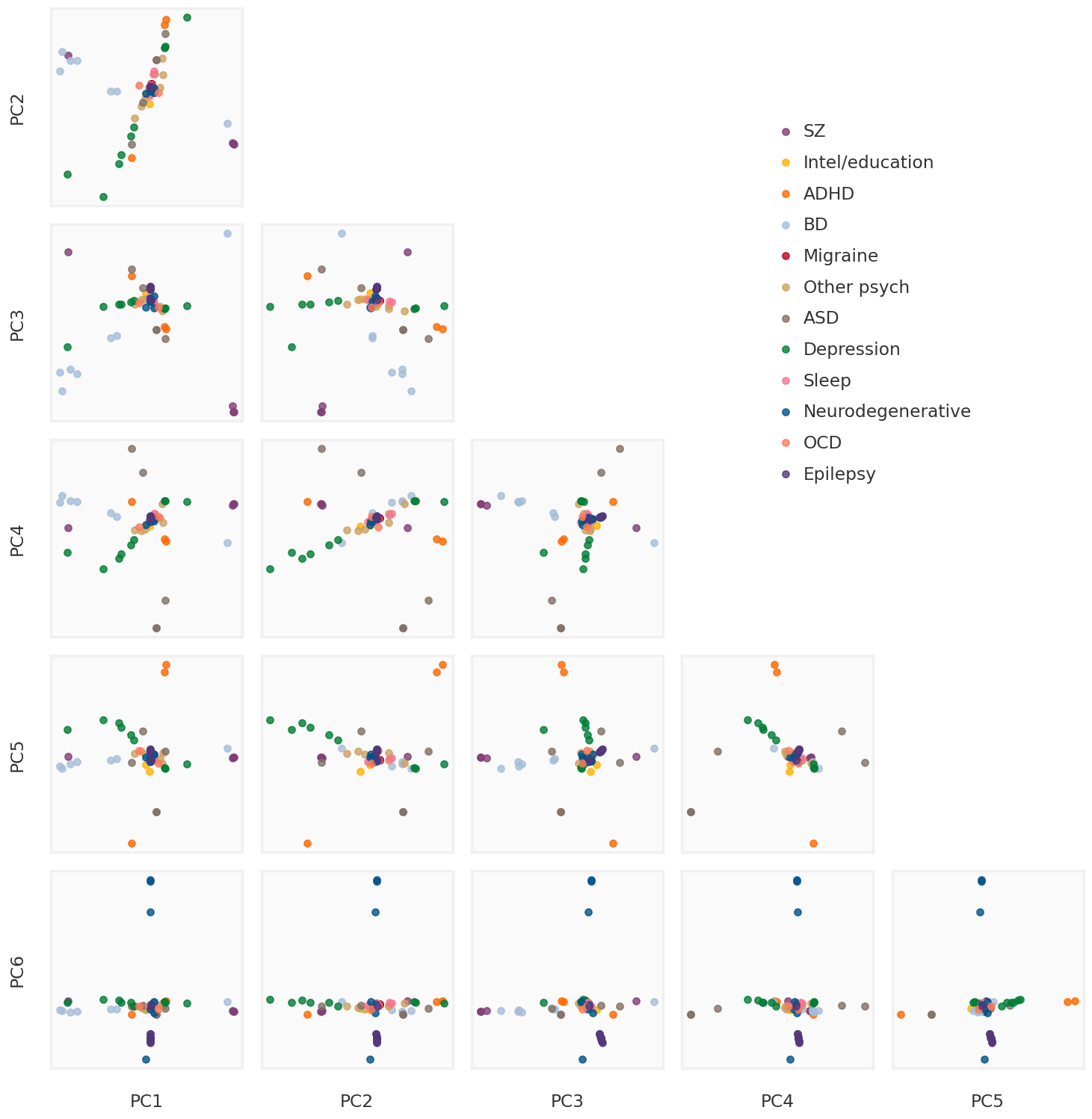
def soft_thresholding(y: np.ndarray, mu: float ):""" Soft thresholding operator as explained in Section 6.5.2 of https://web.stanford.edu/~boyd/papers/pdf/prox_algs.pdf Solves the following problem: argmin_x (1/2)*||x-y||_F^2 + lmb*||x||_1 Parameters ---------- y : np.ndarray Target vector/matrix lmb : float Penalty parameter Returns ------- x : np.ndarray argmin solution """ return np.sign(y) * np.clip(np.abs (y) - mu, a_min= 0 , a_max= None )def svd_shrinkage(y: np.ndarray, tau: float ):""" SVD shrinakge operator as explained in Theorem 2.1 of https://statweb.stanford.edu/~candes/papers/SVT.pdf Solves the following problem: argmin_x (1/2)*||x-y||_F^2 + tau*||x||_* Parameters ---------- y : np.ndarray Target vector/matrix tau : float Penalty parameter Returns ------- x : np.ndarray argmin solution """ = np.linalg.svd(y, full_matrices= False )= soft_thresholding(s, tau)return U.dot(np.diag(s_t)).dot(Vh)class RobustPCA:""" Solves robust PCA using Inexact ALM as explained in Algorithm 5 of https://arxiv.org/pdf/1009.5055.pdf Parameters ---------- lmb: penalty on sparse errors mu_0: initial lagrangian penalty rho: learning rate tau: mu update criterion parameter max_iter: max number of iterations for the algorithm to run tol_rel: relative tolerance """ def __init__ (self , lmb: float , mu_0: float = 1e-5 , rho: float = 2 , tau: float = 10 , int = 10 , tol_rel: float = 1e-3 ):assert mu_0 > 0 assert lmb > 0 assert rho > 1 assert tau > 1 assert max_iter > 0 assert tol_rel > 0 self .mu_0_ = mu_0self .lmb_ = lmbself .rho_ = rhoself .tau_ = tauself .max_iter_ = max_iterself .tol_rel_ = tol_reldef fit(self , X: np.ndarray):""" Fits robust PCA to X and returns the low-rank and sparse components Parameters ---------- X: Original data matrix Returns ------- L: Low rank component of X S: Sparse error component of X """ assert X.ndim == 2 = self .mu_0_= X / self ._J(X, mu)= np.zeros_like(X)= np.empty_like(S)for k in range (self .max_iter_):# Solve argmin_L ||X - (L + S) + Y/mu||_F^2 + (lmb/mu)*||L||_* = svd_shrinkage(X - S + Y/ mu, 1 / mu)# Solve argmin_S ||X - (L + S) + Y/mu||_F^2 + (lmb/mu)*||S||_1 = S.copy()= soft_thresholding(X - L + Y/ mu, self .lmb_/ mu)# Update dual variables Y <- Y + mu * (X - S - L) += mu* (X - S - L)= self ._get_residuals(X, S, L, S_last, mu)# Check stopping cirteria = self ._update_tols(X, L, S, Y)if r < tol_r and h < tol_h:break # Update mu = self ._update_mu(mu, r, h)return L, Sdef _J(self , X: np.ndarray, lmb: float ):""" The function J() required for initialization of dual variables as advised in Section 3.1 of https://people.eecs.berkeley.edu/~yima/matrix-rank/Files/rpca_algorithms.pdf """ return max (np.linalg.norm(X), np.max (np.abs (X))/ lmb)@staticmethod def _get_residuals(X: np.ndarray, S: np.ndarray, L: np.ndarray, S_last: np.ndarray, mu: float ):= np.linalg.norm(X - S - L, ord = "fro" )= mu * np.linalg.norm(S - S_last, ord = "fro" )return primal_residual, dual_residualdef _update_mu(self , mu: float , r: float , h: float ):if r > self .tau_ * h:return mu * self .rho_elif h > self .tau_ * r:return mu / self .rho_else :return mudef _update_tols(self , X, S, L, Y):= self .tol_rel_ * max (np.linalg.norm(X), np.linalg.norm(S), np.linalg.norm(L))= self .tol_rel_ * np.linalg.norm(Y)return tol_primal, tol_dual