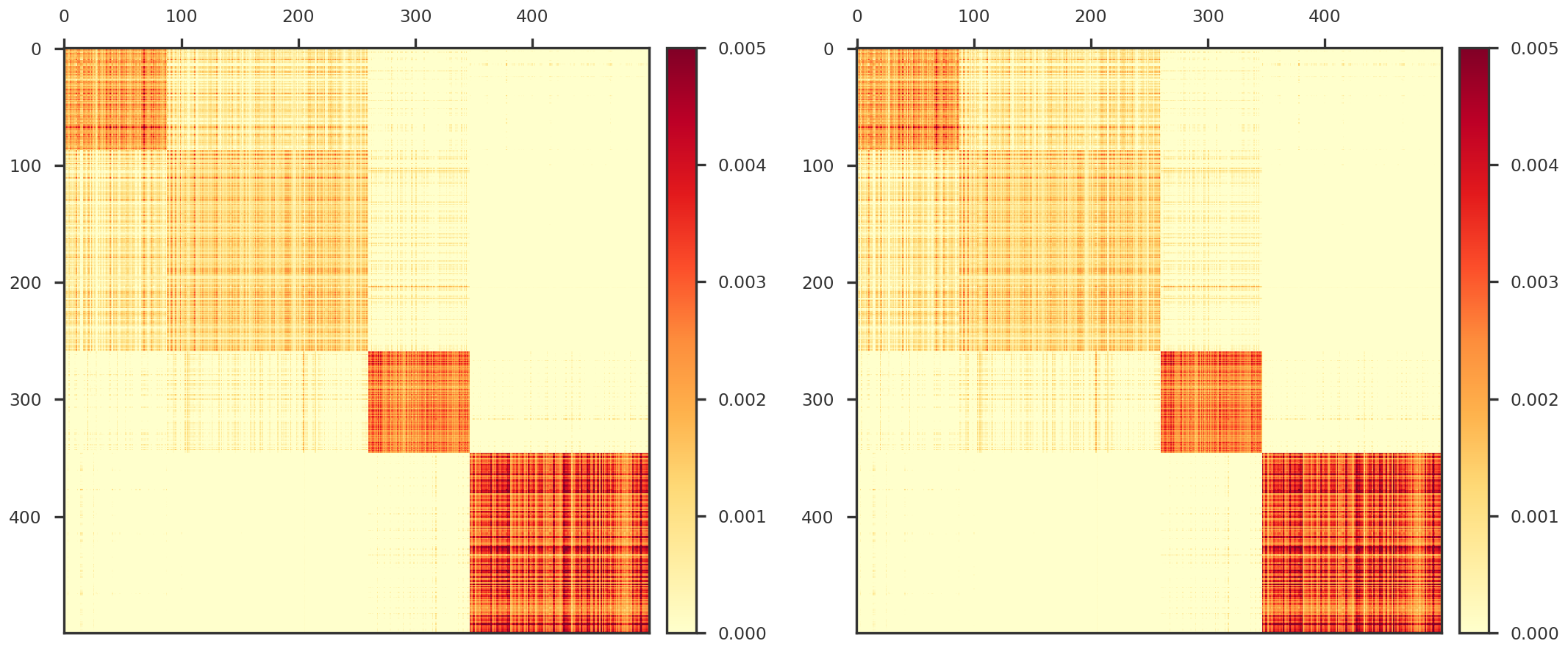
from sklearn.utils.extmath import randomized_svddef nuclear_norm(X):''' Nuclear norm of input matrix ''' return np.sum (np.linalg.svd(X)[1 ])def f_objective(X, Y, W = None , mask = None ):''' Objective function Y is observed, X is estimated W is the weight of each observation. ''' = X if mask is None else X * mask= W if mask is None else W * mask# The * operator can be used as a shorthand for np.multiply on ndarrays. if Wmask is None := 0.5 * np.linalg.norm(Y - Xmask, 'fro' )** 2 else := 0.5 * np.linalg.norm(Wmask * (Y - Xmask), 'fro' )** 2 return f_objdef f_gradient(X, Y, W = None , mask = None ):''' Gradient of the objective function. ''' = X if mask is None else X * mask= W if mask is None else W * maskif Wmask is None := Xmask - Yelse := np.square(Wmask) * (Xmask - Y)return f_graddef linopt_oracle(grad, r = 1.0 , max_iter = 10 , method = 'power' ):''' Linear optimization oracle, where the feasible region is a nuclear norm ball for some r ''' if method == 'power' := singular_vectors_power_method(grad, max_iter = max_iter)elif method == 'randomized' := singular_vectors_randomized_method(grad, max_iter = max_iter)= - r * U1 @ V1_Treturn Sdef singular_vectors_randomized_method(X, max_iter = 10 ):= randomized_svd(X, n_components = 1 , n_iter = max_iter,= 'none' ,= 0 )return u, vhdef singular_vectors_power_method(X, max_iter = 10 ):''' Power method. Computes approximate top left and right singular vector. Parameters: ----------- X : array {m, n}, input matrix max_iter : integer, optional number of steps Returns: -------- u, v : (n, 1), (p, 1) two arrays representing approximate top left and right singular vectors. ''' = X.shape= np.random.normal(0 , 1 , n)/= np.linalg.norm(u)= X.T.dot(u)/= np.linalg.norm(v)for _ in range (max_iter): = X.dot(v)/= np.linalg.norm(u)= X.T.dot(u)/= np.linalg.norm(v) return u.reshape(- 1 , 1 ), v.reshape(1 , - 1 )def do_step_size(dg, D, W = None , old_step = None ):if W is None := np.linalg.norm(D, 'fro' )** 2 else := np.linalg.norm(W * D, 'fro' )** 2 = dg / denom= min (step_size, 1.0 )if step_size < 0 :print ("Warning: Step Size is less than 0" )if old_step is not None and old_step > 0 :print ("Using previous step size" )= old_stepelse := 1.0 return step_sizedef frank_wolfe_minimize_step(X, Y, r, istep, W = None , mask = None , old_step = None , svd_iter = None , svd_method = 'power' ):# 1. Gradient for X_(t-1) = f_gradient(X, Y, W = W, mask = mask)# 2. Linear optimization subproblem if svd_iter is None : = 10 + int (istep / 20 )= min (svd_iter, 25 )= linopt_oracle(G, r, max_iter = svd_iter, method = svd_method)# 3. Define D = X - S# 4. Duality gap = np.trace(D.T @ G)# 5. Step size = do_step_size(dg, D, W = W, old_step = old_step)# 6. Update = X - step * Dreturn Xnew, G, dg, stepdef frank_wolfe_minimize(Y, r, X0 = None ,= None ,= None ,= 1000 ,= None ,= 'power' ,= 1e-4 , step_tol = 1e-3 , rel_tol = 1e-8 ,= True ,= False , debug_step = 10 ):# Step 0 = np.zeros_like(Y) if X0 is None else X0.copy()= np.inf= 1.0 if return_all:= [dg]= [f_objective(old_X, Y, W = weight, mask = mask)]= [1 ]# Steps 1, ..., max_iter for istep in range (max_iter):= \ = weight, mask = mask, old_step = step, svd_iter = svd_iter, svd_method = svd_method)= f_objective(X, Y, W = weight, mask = mask)if return_all:if debug:if (istep % debug_step == 0 ):print (f"Iteration { istep} . Step size { step:.3f} . Duality Gap { dg:g} " )# Stopping criteria # duality gap if np.abs (dg) <= tol:break # step size if step > 0 and step <= step_tol:break # relative tolerance of objective function = np.abs ((f_obj - fx_list[- 2 ]) / f_obj)if f_rel <= rel_tol:break = X.copy()if return_all:return X, dg_list, fx_list, st_listelse :return X